For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRSTTKWSTTDIPDQTGRIAIVTGANTGLGLETAKALAAHGAHVVLAVRNAEKGKAAAEAITAAHSNADV TLQSLDLSSLESVRRASDELKGRYDKIDLLINNAGVMWTEKSSTADGFELQFGTNHLGHYALTGLLLERL LPVEGSRVVTVSSIGHRIRADIHFDDLQWERDYDRVAAYGQSKLANLLFTYELQRRLAGTNTVALAAHPG GSNTELARNSPLWVRAVFDVVAPVLVQGADMGALPTLRAATDPAALGGQYYGPDGFMEQRGNPKVVASSE QSYNLDLQRRLWSVSEELTDVVFPVK*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
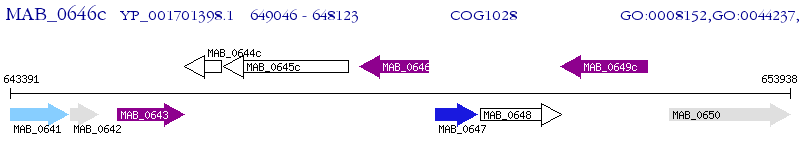
| M. abscessus ATCC 19977 | MAB_0646c | - | - | 100% (307) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0643 | - | e-108 | 68.09% (304) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1144c | - | 5e-62 | 45.95% (309) | short-chain dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0069 | - | 1e-120 | 72.52% (302) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0143 | - | 1e-109 | 67.22% (302) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0068 | - | 1e-120 | 72.19% (302) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 1e-119 | 70.10% (301) | short chain dehydrogenase |
| M. marinum M | MMAR_0757 | - | 1e-124 | 71.66% (307) | dehydrogenase/reductase |
| M. avium 104 | MAV_4710 | - | 1e-117 | 70.86% (302) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0876 | - | 1e-122 | 74.42% (301) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4179 | - | 1e-110 | 65.13% (304) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1391 | - | 1e-124 | 71.34% (307) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0762 | - | 1e-112 | 67.88% (302) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0069|M.bovis_AF2122/97 -------MTKWT---AADIPDQTGRTAVITGANTGLGFETAAALAAHGAH
Rv0068|M.tuberculosis_H37Rv -------MTKWT---AADIPDQTGRTAVITGANTGLGFETAAALAAHGAH
MSMEG_0876|M.smegmatis_MC2_155 -------MTKWT---TARIPDQTGRVAIVTGANTGLGLETAKALAAKGAH
MLBr_00315|M.leprae_Br4923 -------MAKWT---TADIPDQTGRVAVITGANTGLGYQTALALAEHGAH
MAB_0646c|M.abscessus_ATCC_199 ---MSRSTTKWS---TTDIPDQTGRIAIVTGANTGLGLETAKALAAHGAH
MMAR_0757|M.marinum_M MSANGKAKSHWS---ASDIPDQSGRVVVVTGANTGLGYHTAEALAGRGAH
MUL_1391|M.ulcerans_Agy99 MSAKGKAKSHWS---ASDIPDQSGRVVVVTGANTGLGYHTAEALADRGAH
MAV_4710|M.avium_104 MAPHDKRHRDWS---EADVGDQSGRVVVITGANTGIGYETAAVLAHRGAH
TH_4179|M.thermoresistible__bu ------MTGKWT---ARDVPDQTGRTAVITGANTGLGFETAKVLAEKGAH
Mflv_0143|M.gilvum_PYR-GCK -------MTDWTNWNVADIPDQTGRTAVITGANTGLGFETAKALAEKGAR
Mvan_0762|M.vanbaalenii_PYR-1 -------MTDWT---TADIPDQTGRTAVITGANTGLGFETAKALAAGGAR
.*: : **:** .::******:* .** .** **:
Mb0069|M.bovis_AF2122/97 VVLAVRNLDKGKQAAARITEATPGAEVELQELDLTSLASVRAAAAQLKSD
Rv0068|M.tuberculosis_H37Rv VVLAVRNLDKGKQAAARITEATPGAEVELQELDLTSLASVRAAAAQLKSD
MSMEG_0876|M.smegmatis_MC2_155 VVLAVRNLDKGKAAVDWIARSAPTADLELQQLDLGSLASVRAAADDLKGK
MLBr_00315|M.leprae_Br4923 VVLAVRNLDKGKDAAARITATSAQNNVALQELDLASLESVRAAAKQLRSD
MAB_0646c|M.abscessus_ATCC_199 VVLAVRNAEKGKAAAEAITAAHSNADVTLQSLDLSSLESVRRASDELKGR
MMAR_0757|M.marinum_M VVLAVRNPEKGNAAVAQIVAAKPQADVTLQALDLSSLDSVRSAADALRSA
MUL_1391|M.ulcerans_Agy99 VVLAVRNPEKGNAAVAQIVAAKPQADVTLQALDLSSLDSVRSAADALRSA
MAV_4710|M.avium_104 VVLAVRDLEKGNAALSRIVAASPNADVTLQQLDLASLASVRSAAEALRAA
TH_4179|M.thermoresistible__bu VVLAVRDPDKGRRAADRITAAAPHADVTVRQLDLTSLDNIRRAADDLRAG
Mflv_0143|M.gilvum_PYR-GCK VVIAVRNTDKGAQAAARIRG-----DVDVQELDLTSLSSIRTAAEALKAR
Mvan_0762|M.vanbaalenii_PYR-1 VVLAVRDTEKGAQAAAKMAG-----DVDVQQLDLTSLASIRSAADALKSR
**:***: :** * : :: :: *** ** .:* *: *:.
Mb0069|M.bovis_AF2122/97 HQRIDLLINNAGVMYTPRQTTADGFEMQFGTNHLGHFALTGLLIDRLLPV
Rv0068|M.tuberculosis_H37Rv HQRIDLLINNAGVMYTPRQTTADGFEMQFGTNHLGHFALTGLLIDRLLPV
MSMEG_0876|M.smegmatis_MC2_155 FDRIDLLVNNAGVMWPPRQTTADGFELQFGTNHLGHFALTGLLLDRMLTV
MLBr_00315|M.leprae_Br4923 YDHIDLLINNAGVMWTPKSTTKDGFELQFGTNHLGHFAFTGLLLDRLLPI
MAB_0646c|M.abscessus_ATCC_199 YDKIDLLINNAGVMWTEKSSTADGFELQFGTNHLGHYALTGLLLERLLPV
MMAR_0757|M.marinum_M YPRIDLLINNAGVMWTPKQVTKDGFEMQFGTNHLGHFALTGLLLDHLLPV
MUL_1391|M.ulcerans_Agy99 YPRIDLLINNAGVMWTPKQVTKDGFEMQFGTNHLGHFALTGLLLDHLLPV
MAV_4710|M.avium_104 YPRIDLLINNAGVMWTPKQVTEDGFELQFGTNHLGHFALTGLLLDHLLGV
TH_4179|M.thermoresistible__bu YPRIDLLINNAGVMYPPRQTTRDGFELQFGTNHLGHFALTGQLLDNILPV
Mflv_0143|M.gilvum_PYR-GCK FDKIDLLINNAGVMTTPKGTTADGFELQFGTNHLGHFALTGLLFDNILDI
Mvan_0762|M.vanbaalenii_PYR-1 FDHIDLLINNAGVMTTPKGTTADGFELQFGTNHLGHFAFTGLLLDKLLDV
. :****:****** . : * ****:*********:*:** *::.:* :
Mb0069|M.bovis_AF2122/97 AGSRVVTISSVGHRIRAAIHFDDLQWERRYRRVAAYGQAKLANLLFTYEL
Rv0068|M.tuberculosis_H37Rv AGSRVVTISSVGHRIRAAIHFDDLQWERRYRRVAAYGQAKLANLLFTYEL
MSMEG_0876|M.smegmatis_MC2_155 PGSRVVTVSSQGHRILAAIHFDDLQWERRYNRVAAYGQSKLANLLFTYEL
MLBr_00315|M.leprae_Br4923 VGSRVITVSSLSHRLFADIHFNDLQWECNYNRVAAYGQSKLANLLFTYEL
MAB_0646c|M.abscessus_ATCC_199 EGSRVVTVSSIGHRIRADIHFDDLQWERDYDRVAAYGQSKLANLLFTYEL
MMAR_0757|M.marinum_M PGSRVITVSSLGHRIRAAIHFDDLQWERSYNRVAAYGQSKLANLLFTYEL
MUL_1391|M.ulcerans_Agy99 PGSRVITVSSLGHRIRAAIHFDDLQWERSYNRVAAYGQSKLANLLFTYEL
MAV_4710|M.avium_104 RDSRVVTVSSLGHRLRAAIHFDDLHWERRYDRVAAYGQSKLANLLFTYEL
TH_4179|M.thermoresistible__bu DGSRVVTVASIAHRNMADIHFDDLQWERGYHRVAAYGQSKLANLMFAYEL
Mflv_0143|M.gilvum_PYR-GCK PGSRIVTVSSNGHKMGGAIHWDDLQWERSYNRMGAYTQSKLANLLFTYEL
Mvan_0762|M.vanbaalenii_PYR-1 DGARIVTVSSNGHKMGGAIHWDDLQWERSYSRMGAYSQSKLANLLFTYEL
.:*::*::* .*: . **::**:** * *:.** *:*****:*:***
Mb0069|M.bovis_AF2122/97 QRRLAPG--GTTIAVASHPGVSNTELVRNMPRPLV-AVAAILAP-LMQDA
Rv0068|M.tuberculosis_H37Rv QRRLAPG--GTTIAVASHPGVSNTEVVRNMPRPLV-AVAAILAP-LMQDA
MSMEG_0876|M.smegmatis_MC2_155 QRRLT-G--HQTTALAAHPGASNTELARHLPG----ALERLVTP-LAQDA
MLBr_00315|M.leprae_Br4923 QRRLATR--QTTIAVAAHPGGSRTELTRTLPALIA-PIFSVAELFLTQDA
MAB_0646c|M.abscessus_ATCC_199 QRRLA-G--TNTVALAAHPGGSNTELARNSPLWVR-AVFDVVAPVLVQGA
MMAR_0757|M.marinum_M QRRLAADSQAATIAVAAHPGGSNTELARNLPRMLV-PLANILGPALFQSA
MUL_1391|M.ulcerans_Agy99 QRRLAADSQAATIAVAAHPGDSNTELARNLPRMLV-PLANILGPALFQSA
MAV_4710|M.avium_104 QRRLAAAPDAKTIAVAAHPGGSNTELARHLPGIFR-PVQAVLGPVLFQSP
TH_4179|M.thermoresistible__bu QRRLSAK-NAPTISVAAHPGVSNTELTRYIPGARL-PGVSLLAGLLTNSP
Mflv_0143|M.gilvum_PYR-GCK QRRLAPR--GKTIAVAAHPGTSTTELARNLPRPVERAFLAAAPVLFAQTA
Mvan_0762|M.vanbaalenii_PYR-1 QRRLAPR--GKTIAVAAHPGTSSTELGRNLPAALQPALNRLAP-LLAQSP
****: * ::*:*** * **: * * . : : .
Mb0069|M.bovis_AF2122/97 ELGALPTLRAATDPAVRGGQYFGPDGFGEIRGYPKVVASSAQSHDEQLQR
Rv0068|M.tuberculosis_H37Rv ELGALPTLRAATDPAVRGGQYFGPDGFGEIRGYPKVVASSAQSHDEQLQR
MSMEG_0876|M.smegmatis_MC2_155 ALGALPTLRAATDPGALGGQYFGPDGIGETRGYPKVVASSAQSHDADLQR
MLBr_00315|M.leprae_Br4923 ATGALPTLRAATDAAVLGGQYFGPDGFAEIRGHPKVVASNGKSHDVDRQL
MAB_0646c|M.abscessus_ATCC_199 DMGALPTLRAATDPAALGGQYYGPDGFMEQRGNPKVVASSEQSYNLDLQR
MMAR_0757|M.marinum_M QMGALPTLRAATDPSVAGGQYYGPDGFAEQRGHPKIVQSSAQSHDEDLQR
MUL_1391|M.ulcerans_Agy99 QMGALPTLRTATDPSAAGGQYYGPDGFAEQRGHPKIVQSSAQSHDEDLQR
MAV_4710|M.avium_104 AMGALPTLRAATDPAVQGAQYYGPDGFLEQRGRPKLVESSAQSHDEQLQR
TH_4179|M.thermoresistible__bu AVGALATLRAATDPEVKGGQYYGPDGFQEIRGHPVLVGSSAKSRDEDIQR
Mflv_0143|M.gilvum_PYR-GCK DRGALPTLRAAADPGVLGGQYYGPDGLGQQRGAPVVVASSAQSYDVDQQR
Mvan_0762|M.vanbaalenii_PYR-1 AAGALPTLRAATDPSVLGGQYYGPDGIGQQRGNPVVVASSNQSYDMALQR
***.***:*:*. . *.**:****: : ** * :* *. :* : *
Mb0069|M.bovis_AF2122/97 RLWAVSEELTGVVYPVG------
Rv0068|M.tuberculosis_H37Rv RLWAVSEELTGVVYPVG------
MSMEG_0876|M.smegmatis_MC2_155 RLWAVSEELTGVTFPVGEALQQA
MLBr_00315|M.leprae_Br4923 RLWAVSEELTGVVYPVG------
MAB_0646c|M.abscessus_ATCC_199 RLWSVSEELTDVVFPVK------
MMAR_0757|M.marinum_M RLWTVSEELTGVSFPV-------
MUL_1391|M.ulcerans_Agy99 RLWTVSEELTGVSFPV-------
MAV_4710|M.avium_104 RLWAVSEELTGVHFPV-------
TH_4179|M.thermoresistible__bu RLWTVSEELTGVTFPV-------
Mflv_0143|M.gilvum_PYR-GCK RLWEISEELTKVTFPDL------
Mvan_0762|M.vanbaalenii_PYR-1 RLWTVSEELTGVTFPVPVLS---
*** :***** * :*