For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRSPTPRRRATLASLAAELKVSRTTVSNAYNRPDQLSAELRERVLSTAERLGYPGPDPVARSLRTRRAGA VGLVITEPLNYSFSDPAALDFVAGLAESCEAVGQGLLLVAVGPNRSVADGSAAVLAAGVDGFVVYSASDD DPYLPAVLQRHLPVVVVDQPRDVAGTSRVCIDDRAAMRRLAEHVIELGHREIGLLTMRLGRDRPHGGSTP VLADPERLRTPHFHVQGERIRGVYDAMTAAGLDPGSLTVVESYEHLPSSGGAGADVALDANPRITALMCT ADVLALSAMDHLRAKGIYVPGQMTVTGFDGVPEARRRGLTTVVQPSVEKGRRAGELLHHPPRSGLPVIEV LDTEMVRGRTSGPPA*
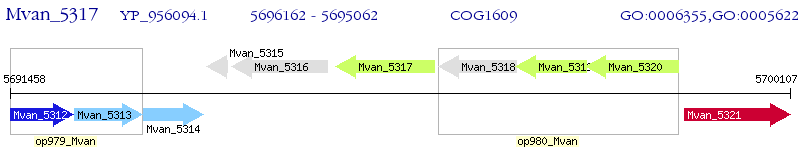
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5317 | - | - | 100% (366) | periplasmic binding protein/LacI transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2545 | - | 2e-25 | 29.97% (367) | periplasmic binding protein/LacI transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0385 | - | 4e-20 | 28.65% (370) | periplasmic binding protein/LacI transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3606c | - | 1e-159 | 79.06% (363) | transcriptional regulatory protein LacI-family |
| M. gilvum PYR-GCK | Mflv_1457 | - | 0.0 | 90.98% (366) | periplasmic binding protein/LacI transcriptional regulator |
| M. tuberculosis H37Rv | Rv3575c | - | 1e-159 | 79.06% (363) | transcriptional regulatory protein LacI-family |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2023c | - | 1e-09 | 27.92% (197) | LacI family transcriptional regulator |
| M. marinum M | MMAR_5070 | - | 1e-159 | 77.69% (363) | PurR family transcriptional regulator |
| M. avium 104 | MAV_0585 | - | 1e-159 | 78.51% (363) | transcriptional regulator, LacI family protein |
| M. smegmatis MC2 155 | MSMEG_6044 | - | 1e-173 | 81.97% (366) | ligand-binding /sugar binding domain-containing protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4146 | - | 1e-159 | 77.41% (363) | PurR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5317|M.vanbaalenii_PYR-1 MPRSPTPRRRATLASLAAELKVSRTTVSNAYNRPDQLSAELRERVLSTAE
Mflv_1457|M.gilvum_PYR-GCK MSRSPTPRRRATLASLAAELKVSRTTVSNAYNRPDQLSAELRERVLSTAK
MSMEG_6044|M.smegmatis_MC2_155 MPRSPHPPRRATLASLAAELKVSRTTISNAYNRPDQLSADLRERIFDAAK
Mb3606c|M.bovis_AF2122/97 --MSPTPRRRATLASLAAELKVSRTTVSNAFNRPDQLSADLRERVLATAK
Rv3575c|M.tuberculosis_H37Rv --MSPTPRRRATLASLAAELKVSRTTVSNAFNRPDQLSADLRERVLATAK
MMAR_5070|M.marinum_M --MSPTPRRRATLASLAAELKVSRSTISNAFNRPDQLSAELRERVLATAK
MUL_4146|M.ulcerans_Agy99 --MSPTPRRRATLASLAAELKVSRSTISNAFNRPDQLSVELRERVLATAK
MAV_0585|M.avium_104 --MSPTPRRRATLASLADELKVSRTTVSNAFNRPDQLSADLRERVLATAK
MAB_2023c|M.abscessus_ATCC_199 -------------------MGLSKTTVADALGGSGRVSEATREKTRLVAE
: :*::*:::* . ..::* **: .*:
Mvan_5317|M.vanbaalenii_PYR-1 RLGYPGPDPVARSLRTRRAGAVGLVITEPLNYSFSDPAALDFVAGLAESC
Mflv_1457|M.gilvum_PYR-GCK RLGYPGPDPVARSLRTRRAGAVGLVITEPLNYSFSDPAALDFVAGLAESC
MSMEG_6044|M.smegmatis_MC2_155 RLGYPGPDPVARSLRTRKAGAVGLMITEPLDYSFSDPAALDFVAGLAESC
Mb3606c|M.bovis_AF2122/97 RLGYAGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVAGVAQSC
Rv3575c|M.tuberculosis_H37Rv RLGYAGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVAGVAQSC
MMAR_5070|M.marinum_M RMGYPGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVTGVAQSC
MUL_4146|M.ulcerans_Agy99 RMGYPGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVTGVAQSC
MAV_0585|M.avium_104 RLGYPGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVTGVAQSC
MAB_2023c|M.abscessus_ATCC_199 RMGYVS-NRAARLLRRNETGALGLYIPGDVRNLS---FYMELAFGAADES
*:** . : .** ** ..:**:** :. : ::. * *:..
Mvan_5317|M.vanbaalenii_PYR-1 EAVGQGLLLVAVGPNRSVADGSAAVLAAGVDGFVVYSASDDDPYLPAVLQ
Mflv_1457|M.gilvum_PYR-GCK EAVGQGLLLVAVGPNRSVAEGSAAVLAAGVDGFVVYSASDDDPYLPAVME
MSMEG_6044|M.smegmatis_MC2_155 EEAGQGLLLVAVGPNRTFEEGSNSVLAAGVDGFVVYSASDDDPYLPVLQQ
Mb3606c|M.bovis_AF2122/97 EELGQGLQLVSVGSSRSLADGTAAVLGAGVDGFVVYSVGDDDPYLQVVLQ
Rv3575c|M.tuberculosis_H37Rv EELGQGLQLVSVGSSRSLADGTAAVLGAGVDGFVVYSVGDDDPYLQVVLQ
MMAR_5070|M.marinum_M EELGQGLLLVAVGASRSLEDGTAAVLNAGVDGFVVYSVRDDDPYLQVVLQ
MUL_4146|M.ulcerans_Agy99 EELGQGLLLVAVGASRSLEDGTAAVLNAGVDGFVVYSVRDDDPYLQVVLQ
MAV_0585|M.avium_104 EALGQGLLLVAVGPSRSLDEGTAAVLSAGVDGFVVYSVCDDDPYLQAVLQ
MAB_2023c|M.abscessus_ATCC_199 AALGLDLTLIGR-----HLTGENRSRPPQVDGIIAVDPVPGDAALESLLT
* .* *:. * . ***::. . .*. * :
Mvan_5317|M.vanbaalenii_PYR-1 RHLPVVVVDQPRDVAG---TSRVCIDDRAAMRRLAEHVIELGHREIGLLT
Mflv_1457|M.gilvum_PYR-GCK RHLPVVVVDQPRDVPG---ASQVRIDDRAAMRSLAEHVIDLGHREIGLLT
MSMEG_6044|M.smegmatis_MC2_155 RQLPMVVVDQPKDVPG---VSRISIDDRGAMRELAEYVLELGHRDIGLLT
Mb3606c|M.bovis_AF2122/97 RRLPVVVVDQPKDLSG---VSRVGIDDRAAMRELAGYVLGLGHRELGLLT
Rv3575c|M.tuberculosis_H37Rv RRLPVVVVDQPKDLSG---VSRVGIDDRAAMRELAGYVLGLGHRELGLLT
MMAR_5070|M.marinum_M RRLPVVVVDQPKELAG---VSRVGIDDRAAMRKIADYVLGLGHRQIGLLT
MUL_4146|M.ulcerans_Agy99 RRLPVVVVDQPKELAG---VSRVGIDDRAAMRKIADYVLSLGHRQIGLLT
MAV_0585|M.avium_104 RRLPVVVVDQPKGLTG---VSRVGIDDRAAMRELADYVLGLGHREIGLLT
MAB_2023c|M.abscessus_ATCC_199 SNVPIVTVGRILDKDVSHHAAEIEIDHYRMVTLLLDAIRAAGGKRPALAA
.:*:*.*.: .:.: **. : : : * : .* :
Mvan_5317|M.vanbaalenii_PYR-1 MRLGRDRPHGGSTPVLADPERLRTPHFHVQGERIRGVYDAMTAAGLDPGS
Mflv_1457|M.gilvum_PYR-GCK MRLGRDWPHG-PGPALADPERVRAPNFHVQCERIHGVYDAMTAAGLDPAS
MSMEG_6044|M.smegmatis_MC2_155 MRLGRDWPHGDPRPALADPERVLSPHFHVQRERIHGVYDAMSAAGLDPSS
Mb3606c|M.bovis_AF2122/97 MRLGRDRRQD-----LVDAERLRSPTFDVQRERIVGVWEAMTAAGVDPDS
Rv3575c|M.tuberculosis_H37Rv MRLGRDRRQD-----LVDAERLRSPTFDVQRERIVGVWEAMTAAGVDPDS
MMAR_5070|M.marinum_M MRLGRDRRQD-----LVDAERLKSPTFDVQRERITGVWEAMTAAGVDPES
MUL_4146|M.ulcerans_Agy99 MRLGRDRRQD-----LVDAERLKSPTFDVQRERITGVWEAMTAAGVDPES
MAV_0585|M.avium_104 MRLGRDRRQG-----LVDAERLRSPAFDVQRERITGVWEAMTAAGVDPDS
MAB_2023c|M.abscessus_ATCC_199 LDQGAD----------------SSYVFDVSR----GYQEWCAKAGVVPLA
: * * : *.*. * : : **: * :
Mvan_5317|M.vanbaalenii_PYR-1 LTVVESYEHLPSSGGAGADVALDANPRITALMCTADVLALSAMDHLRAKG
Mflv_1457|M.gilvum_PYR-GCK LTLVESYEHLPTSGGAGADIALEANPRITALMCTADVLALSAMDHLRAKG
MSMEG_6044|M.smegmatis_MC2_155 LTVVESYEHLPTSGGAAAEVALEVNPRITALMCTADVLALSAMDHLRSRG
Mb3606c|M.bovis_AF2122/97 LTVVESYEHLPTSGGTAAKVALQANPRLTALMCTADILALSAMDYLRAHG
Rv3575c|M.tuberculosis_H37Rv LTVVESYEHLPTSGGTAAKVALQANPRLTALMCTADILALSAMDYLRAHG
MMAR_5070|M.marinum_M LTVVESYEHLPTSGGDAAKVALEANPRITALMCTADILALSAMDYLRGHG
MUL_4146|M.ulcerans_Agy99 LTVVESYEHLPTSGGDAAKVALEANPRITALMCTADILALSAMDYLRGHG
MAV_0585|M.avium_104 LTVVESYEHLPESGGAAAKVALEANPRITALMCTADILALSAMDYLRAHG
MAB_2023c|M.abscessus_ATCC_199 TRLSATPTDVELAQAIRGIVEHQG---IDALVVGAQGLAARSLPVLQSLG
: : .: : . . : : : **: *: ** :: *:. *
Mvan_5317|M.vanbaalenii_PYR-1 IYVPGQMTVTGFDG--VPEARRRGLTTVVQPSVEKGRRAGELLH--HPPR
Mflv_1457|M.gilvum_PYR-GCK IYVPGQMTVTGFDG--VPEARQRGLTTVAQPGVEKGRRAGELLH--NPPR
MSMEG_6044|M.smegmatis_MC2_155 IYVPGHMTVTGFDG--VRDALRRGLTTVKQPSMEKGRRAGHLLH--NPPG
Mb3606c|M.bovis_AF2122/97 IYVPGQMTVTGFDG--VPEALSRGLTTVAQPSLHKGHRAGELLL--KPPR
Rv3575c|M.tuberculosis_H37Rv IYVPGQMTVTGFDG--VPEALSRGLTTVAQPSLHKGHRAGELLL--KPPR
MMAR_5070|M.marinum_M IYVPGQMTVTGFDG--VPEAISRGLTTVSQPSLQKGRRAGELLQ--NPPR
MUL_4146|M.ulcerans_Agy99 IYVPGQMTVTGFDG--VPEAISRGLTTVSQPSLQKGRRAGELLQ--NPPR
MAV_0585|M.avium_104 VYVPGQLTVTGFDG--VPDAISRGLTTVSQSSLHKGRRAGELLL--KPAR
MAB_2023c|M.abscessus_ATCC_199 HEVGSNFHLGTLVGDPATELTNPDIHAVDLRPREFGHQAVTFLHDIIAGR
* .:: : : * . : .: :* . *::* :* .
Mvan_5317|M.vanbaalenii_PYR-1 SGLPVIEVLDTEMVRGRTSGPPA-------
Mflv_1457|M.gilvum_PYR-GCK SGLPVVEVLDTEVIRGRTSGPPA-------
MSMEG_6044|M.smegmatis_MC2_155 SGLPVIDVLETEVVRGRTSGPPA-------
Mb3606c|M.bovis_AF2122/97 SGLPVIEVLDTELVRGRTAGPPA-------
Rv3575c|M.tuberculosis_H37Rv SGLPVIEVLDTELVRGRTAGPPA-------
MMAR_5070|M.marinum_M SGLPVVELLDTELIRGRTAGPPA-------
MUL_4146|M.ulcerans_Agy99 SGLPVVELLDTELIRGRTAGPPA-------
MAV_0585|M.avium_104 SGLPVVELLDTELIRGRTAGPPA-------
MAB_2023c|M.abscessus_ATCC_199 ASANDRRTHQARLKIAGVSSPPAIRRAMSD
:. ::.: . .:.***