For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPRRKAIILVSCCLSLLIVSMDATIVNVAIPSIRDDLSATPAQMQWVVDVYTLVLASLLMLSGATGDRFG RRRVFQIGLAVFALGSLACSLAPTIDALIGARLLQGIGGSMLNPVALSIISQIFTQPVERARALGIWGGV TGISMAAGPIVGGLLIDTIGWRSVFWINLPICAAAILLTAIFVPESKSQTMRNVDPIGQGLAIMFLFGTV FTLIEGPVLGWTNPRVVVAAVVVALALVAFLRFESRRHDPFLDLRFFRSIPFTTATVIAISAFAAWGAFL FLMSLYLQGERGFSAMHTGLIYLPIAIGALLFSPLSGRLVGRYGARPSLVIAGVTITAAATMLTFLTATT PVWSLLMVFAVFGIGFSMVNAPVTNAAVSGMPLDRAGAASAVTSTSRQVGVSIGVALCGSLAGSAMATGG ADFAAAARPLWFVCMALGLVILGLGLFATSQRALRSAERLAPLVSGAPEPVGGAHVR*
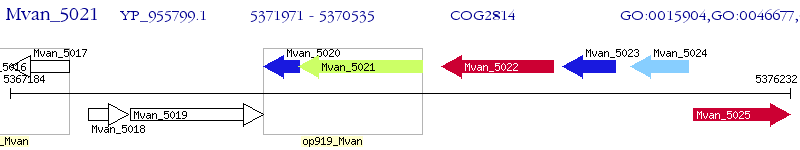
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5021 | - | - | 100% (478) | EmrB/QacA family drug resistance transporter |
| M. vanbaalenii PYR-1 | Mvan_0523 | - | 0.0 | 66.81% (467) | EmrB/QacA family drug resistance transporter |
| M. vanbaalenii PYR-1 | Mvan_1284 | - | 4e-62 | 33.10% (423) | major facilitator superfamily transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2361c | - | 1e-75 | 35.94% (409) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_1730 | - | 0.0 | 88.49% (478) | EmrB/QacA family drug resistance transporter |
| M. tuberculosis H37Rv | Rv2333c | - | 6e-75 | 35.94% (409) | integral membrane transport protein |
| M. leprae Br4923 | MLBr_01562 | - | 4e-50 | 30.73% (397) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_0945 | - | 0.0 | 67.45% (467) | EmrB/QacA family drug resistance transporter |
| M. marinum M | MMAR_1146 | - | 2e-80 | 38.08% (407) | integral membrane transport protein |
| M. avium 104 | MAV_1387 | - | 4e-58 | 34.01% (397) | drug transporter |
| M. smegmatis MC2 155 | MSMEG_5670 | - | 0.0 | 73.43% (478) | efflux membrane protein |
| M. thermoresistible (build 8) | TH_3965 | - | 4e-58 | 34.20% (424) | PUTATIVE drug transporter |
| M. ulcerans Agy99 | MUL_0911 | - | 6e-81 | 36.72% (433) | integral membrane transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5021|M.vanbaalenii_PYR-1 -------------------------------------------------M
Mflv_1730|M.gilvum_PYR-GCK -------------------------------------------------M
MSMEG_5670|M.smegmatis_MC2_155 ------------------------------------------MNETVGAL
MAB_0945|M.abscessus_ATCC_1997 ------------------------------------------MEAAVGAL
MAV_1387|M.avium_104 --------------------------------------------------
TH_3965|M.thermoresistible__bu ----------------------------VTSVDAGRSTMSEVFTAYAPRS
Mb2361c|M.bovis_AF2122/97 --------------------------------------------------
Rv2333c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------MRIQPV
MUL_0911|M.ulcerans_Agy99 --------------------------------------------MRIQPV
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Mvan_5021|M.vanbaalenii_PYR-1 TPRRKAIILVSCCLSLLIVSMDATIVNVAIPSIRDDLSATPAQMQWVVDV
Mflv_1730|M.gilvum_PYR-GCK TPRRKAIILVSCCLSLLIVSMDATIVNVAIPSIRADLSASPAQMQWVVDI
MSMEG_5670|M.smegmatis_MC2_155 SARRKAIILVSCCLSLLIVSMDATIVNVAIPSIRSDLGASPSQLQWVIDV
MAB_0945|M.abscessus_ATCC_1997 SERRKIVILGSCCLSLLIVSMDSTIVNVALPTIRADFGATTSQLQWVIDI
MAV_1387|M.avium_104 -----------MMIGFFMIMVDSTIVAIANPTIMADLHIGYDTVVWVTSA
TH_3965|M.thermoresistible__bu AAANPWHALWAMLIGFFMILVDATIVPVANLEIMADLGADYTAVIWVTSA
Mb2361c|M.bovis_AF2122/97 MNRTQLLTLIATGLGLFMIFLDALIVNVALPDIQRSFAVGEDGLQWVVAS
Rv2333c|M.tuberculosis_H37Rv MNRTQLLTLIATGLGLFMIFLDALIVNVALPDIQRSFAVGEDGLQWVVAS
MMAR_1146|M.marinum_M FRDHPIPALAVICLSVFVISVDATIVNVALPTLSRQLGADTAQLQWIVDA
MUL_0911|M.ulcerans_Agy99 FRDHPILALAVICLSVFVISVDATIVNVALPTLSRELGADTAQLQWIVDA
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
:: :*: :. :* : .: *
Mvan_5021|M.vanbaalenii_PYR-1 YTLVLASLLMLSGATGDRFGRRRVFQIGLAVFALGSLACSLAPTIDALIG
Mflv_1730|M.gilvum_PYR-GCK YTLVLASLLMLSGATGDRFGRRRLFQIGLTVFALGSLACSLAPTIDTLIG
MSMEG_5670|M.smegmatis_MC2_155 YTLVLASLLLLAGATGDRFGRRRTFHLGLTVFAVASLLCSVAPNIETLIA
MAB_0945|M.abscessus_ATCC_1997 YTLGLASLLMLSGAAGDRLGRRKVFHLGLSIFAIGSLLCSVAPTVGLLIA
MAV_1387|M.avium_104 YLLGYAVVLLVAGRLGDRFGTKNLYLIGLAVFTVASVWCGLAGSAAMLIA
TH_3965|M.thermoresistible__bu YLLAYAVPLLIAGRLGDRFGPRTMYLIGLAIFTAASLWCGLAGTIGMLIA
Mb2361c|M.bovis_AF2122/97 YSLGMAVFIMSAATLADLYGRRRWYLIGVSLFTLGSIACGLAPSIAVLTT
Rv2333c|M.tuberculosis_H37Rv YSLGMAVFIMSAATLADLDGRRRWYLIGVSLFTLGSIACGLAPSIAVLTT
MMAR_1146|M.marinum_M YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLIA
MUL_0911|M.ulcerans_Agy99 YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLVA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
* * . :: .. .* * : *: :*: * .. . *
Mvan_5021|M.vanbaalenii_PYR-1 ARLLQGIGGSMLNPVALSIISQIFTQPVERARALGIWGGVTGISMAAGPI
Mflv_1730|M.gilvum_PYR-GCK ARFLQGVGGSMLNPVALSIISQIFTKPVERARALGIWGGVTGISMAAGPI
MSMEG_5670|M.smegmatis_MC2_155 ARLLQAIGGSMLNPVAMSIITQVFTGRVERARAIGVWGSVVGISMALGPL
MAB_0945|M.abscessus_ATCC_1997 ARLLQAVGGSMLNPVALSIISQVFTGRVERARALGFWGAVVGISMALGPI
MAV_1387|M.avium_104 ARVVQGVGAGVMTPQTLSTITRIFPAERR-GVAVSVWSATAGAASLAGPL
TH_3965|M.thermoresistible__bu ARVLQGLGAALLTPQTLSMITRIFPADRR-GVPMGVWGATAGVATLIGPL
Mb2361c|M.bovis_AF2122/97 ARGAQGLGAAAVSVTSLALVSAAFPEAKEKARAIGIWTAIASIGTTTGPT
Rv2333c|M.tuberculosis_H37Rv ARGAQGLGAAAVSVTSLALVSAAFPEAKEKARAIGIWTAIASIGTTTGPT
MMAR_1146|M.marinum_M ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
MUL_0911|M.ulcerans_Agy99 ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
** .:*.. :. :: *. . ..: . .. . *
Mvan_5021|M.vanbaalenii_PYR-1 VGGLLIDTIGWRSVFWINLPICAAAILLTAIFVPESKSQTMRNVDPIGQG
Mflv_1730|M.gilvum_PYR-GCK VGGLLIETVGWRSVFWINLPICAAAIILTAVFVPESKSQTMRNVDPIGQG
MSMEG_5670|M.smegmatis_MC2_155 VGGALIEYVDWRAVFWINLPICALAILLTAIFVRESKSSTMRDVDPVGQG
MAB_0945|M.abscessus_ATCC_1997 VGGLLIQWVGWRAVFWINLPVCVLAFVLTAVFVPETKSATMRDIDPVGQA
MAV_1387|M.avium_104 AGGVLVDGLGWQWIFFVNVPIGVLGLVLAYWLVPVLP-TQSHRFDLVGVG
TH_3965|M.thermoresistible__bu AGGVLVDGLGWQWIFIVNVPIGIAGLVLGARLMPRLP-RQDQGFDVLGMV
Mb2361c|M.bovis_AF2122/97 LGGLLVDQWGWRSIFYVNLPMGALVLFLTLCYVEESCNERARRFDLSGQL
Rv2333c|M.tuberculosis_H37Rv LGGLLVDQWGWRSIFYVNLPMGALVLFLTLCYVEESCNERARRFDLSGQL
MMAR_1146|M.marinum_M TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLM
MUL_0911|M.ulcerans_Agy99 TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLM
MLBr_01562|M.leprae_Br4923 VGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKERMK-LDAAGAI
** * : * * :*:*: : : .* *
Mvan_5021|M.vanbaalenii_PYR-1 LAIMFLFGTVFTLIEGPVLGWTNPRVVVAAVVVALALVAFLRFESRRHDP
Mflv_1730|M.gilvum_PYR-GCK LGILFLFGTVFTLIEGPVLGWTNPRVLAIAAVAVVALIAFLRFESRRHDP
MSMEG_5670|M.smegmatis_MC2_155 LGVAFLFGVVFVLIEGPGMGWTDPRTLAVAATALVAFLAFLRYESRRHDP
MAB_0945|M.abscessus_ATCC_1997 LAVLFLFGIVFALIEGPSLGWATPGLIVVLAVALAAFAVFLRYESRRRHP
MAV_1387|M.avium_104 LSGVGMLLIVFGLQQGQAAHWQPWIWALIVAGVGFVTVFVFWQSVNVREP
TH_3965|M.thermoresistible__bu LSAVGLFLIVFALQEAQTHRWAPWIWGTLAGGAGVMAMFVIWQSANRRHP
Mb2361c|M.bovis_AF2122/97 LFIVAVGALVYAVIEGPQIGWTSVQTIVMLWTAAVGCALFVWLERRSSNP
Rv2333c|M.tuberculosis_H37Rv LFIVAVGALVYAVIEGPQIGWTSVQTIVMLWTAAVGCALFVWLERRSSNP
MMAR_1146|M.marinum_M LSAVGITALVYTIIEAPNWGWTSTRASGGFIAAAIVLVGFALWERRSSHP
MUL_0911|M.ulcerans_Agy99 LSAVGITALVYTIIEAPNWGWTSTRASGGFIAAAIVLVGFALWERRSSHP
MLBr_01562|M.leprae_Br4923 LATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAENP
* *: . . * . . . .*
Mvan_5021|M.vanbaalenii_PYR-1 FLDLRFFRSIPFTTATVIAISAFAAWGAFLFLMSLYLQGERGFSAMHTGL
Mflv_1730|M.gilvum_PYR-GCK FLDLRFFRSIPFTTATINAISAFAAWGAFLFMMSLYLQGERGFSAMQTGL
MSMEG_5670|M.smegmatis_MC2_155 FIDLRFFRSVPFASATLVAVCAFAAYGAFLFMMSLYLQSERGYSAMFTGL
MAB_0945|M.abscessus_ATCC_1997 FIDLRFFRSIPFASATLTAICACMAWSAFLFTMSLYLQGQRHFSAMHTGL
MAV_1387|M.avium_104 LIPLVIFSDRDFSLCNIGVAIISFAATAMMLPLTFYAQAVCGLSPTRSAL
TH_3965|M.thermoresistible__bu LVPLQIFADRNFMLANVGVATIGFVVTAMVLPAMFFAQAVCGLSPTGSAL
Mb2361c|M.bovis_AF2122/97 MMDLTLFRDTSYALAIATICTVFFAVYGMLLLTTQFLQNVRGYTPSVTGL
Rv2333c|M.tuberculosis_H37Rv MMDLTLFRDTSYALAIATICTVFFAVYGMLLLTTQFLQNVRGYTPSVTGL
MMAR_1146|M.marinum_M MLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQTGV
MUL_0911|M.ulcerans_Agy99 MLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQTGV
MLBr_01562|M.leprae_Br4923 VVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALRAGI
.: . .* . . .: . : * :. :.:
Mvan_5021|M.vanbaalenii_PYR-1 IYLPIAIGALLFSPLSGRLVGRYGARPSLVIAG-VTITAAATMLTFLTAT
Mflv_1730|M.gilvum_PYR-GCK IYLPIALGALFCSPLSGRLVGRYGARPSLVAAG-ILITTAATLLTLLTAT
MSMEG_5670|M.smegmatis_MC2_155 IYLPIAIGALIFSPLSGRMVGRFGSRPSLVSAG-VLITAATVMLAQMTAT
MAB_0945|M.abscessus_ATCC_1997 IYLPIAVGALVFSPISGRMVGRFGARPSLLAAG-TLFLTASLLLTRVSEA
MAV_1387|M.avium_104 LIAPMAIANGVFAPFVGKIVDRYHPRPVLGFGFSLLAIALTWLTFEMSPA
TH_3965|M.thermoresistible__bu LIAPMAVASGVLAPMAGRIVDRSHPTPVVGFGFSVVAVGLTWLAVEMAPT
Mb2361c|M.bovis_AF2122/97 MILPFSAAVAIVSPLVGHLVGRIGARVPILAGLCMLMLGLLMLIFSEHRS
Rv2333c|M.tuberculosis_H37Rv MILPFSAAVAIVSPLVGHLVGRIGARVPILAGLCMLMLGLLMLIFSEHRS
MMAR_1146|M.marinum_M RLLPVALSIALAAILGPRLVERVGTTAVVAAGLAVFAAALAWAS-TADAT
MUL_0911|M.ulcerans_Agy99 RLLPVAFSIALAAILGPRLVERVGTTAVVAAGLAVFAAALAWAS-TADAT
MLBr_01562|M.leprae_Br4923 GFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMNRG
*. . . . ::* * . .
Mvan_5021|M.vanbaalenii_PYR-1 TPVWS-LLMVFAVFGIGFSMVNAPVTNAAVSGMPLDRAGAASAVTSTSRQ
Mflv_1730|M.gilvum_PYR-GCK TPVWH-LLIVFAVFGVGFSMVNAPITNAAVSGMPLDRAGAASAVTSTSRQ
MSMEG_5670|M.smegmatis_MC2_155 TPVWL-LLVIFAVFGIGFALVNAPITTAAVSGMPLDRAGAASAVASTSRQ
MAB_0945|M.abscessus_ATCC_1997 TPIWE-LLVTFAIFGAGFAMVNAPITNAAVSGMPLDRAGAASAVTSTSRQ
MAV_1387|M.avium_104 TPIWR-LVLPFFAMGVGMAFVWSPLTATATRNLSAQLAGAGSAVYNSVRQ
TH_3965|M.thermoresistible__bu TPIWR-LVLPFVAVGVGMAFIWSPLATTATRQLPLHLSGAGSGVYNTTRQ
Mb2361c|M.bovis_AF2122/97 SALVL-VGLGLCGS--GVALCLTPITTVAMTAVPAERAGMASGIMSAQRA
Rv2333c|M.tuberculosis_H37Rv SALVL-VGLGLCGS--GVALCLTPITTVAMTAVPAERAGMASGIMSAQRA
MMAR_1146|M.marinum_M TPYTQ-IALQMLLLGGGLGLTTAPATEAIMGSLPADRAGVGSAVNDTTRE
MUL_0911|M.ulcerans_Agy99 TPYTQ-ITLQMLLLGGGLGLTTAPATEAIMGSLPADRAGVGSAVNDTTRE
MLBr_01562|M.leprae_Br4923 VPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALMLQS
. : : *..: * : : . * *.: :
Mvan_5021|M.vanbaalenii_PYR-1 VGVSIGVALCGSLAGS------AMATGGADFAAAARPLWFVCMALGLVIL
Mflv_1730|M.gilvum_PYR-GCK VGVSIGVALCGSVAGA------AMAGSGTDFATAARPLWFVCIALGLVIL
MSMEG_5670|M.smegmatis_MC2_155 VGVSIGVALCGSVAGT------ALAGTGVDFAAASRPLWLICTALGVVIV
MAB_0945|M.abscessus_ATCC_1997 VGVSIGVALCGSIG--------------------TPALWFMCAGLGLLIA
MAV_1387|M.avium_104 LGAVLGSAGMAAFMTW------RIGAEMP-GQPGGGGEDSAGPVLPEFLR
TH_3965|M.thermoresistible__bu IGSVLGSAGMAAFLTG------RLSAELP-FGATAVPAGEQTRVLPEFLH
Mb2361c|M.bovis_AF2122/97 IGSTIGFAVLGSVLAA------WLSATLEPHLERAVPDPVQRHVLAEIII
Rv2333c|M.tuberculosis_H37Rv IGSTIGFAVLGSVLAA------WLSATLEPHLERAVPDPVQRHVLAEIII
MMAR_1146|M.marinum_M LGGTLGVAIAGSVFAS------VYSGQLGSASTLATLPSDVRSAMKGSMA
MUL_0911|M.ulcerans_Agy99 LGGTLGVAIAGSVFAS------VYSGQLGSASALATLPSDVRSAMKGSMA
MLBr_01562|M.leprae_Br4923 LGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGYTYGLLWV
:* : * :.
Mvan_5021|M.vanbaalenii_PYR-1 GLGLFATSQRALRSAERLAPLVSGAPEPVGGAHVR---------------
Mflv_1730|M.gilvum_PYR-GCK AVGFFSTSQRAVRSAERLAPLVAGAPDPVNGAHVR---------------
MSMEG_5670|M.smegmatis_MC2_155 AIGIVSTSPWALRSAERLAPLIGQHHRPAEVAHAR---------------
MAB_0945|M.abscessus_ATCC_1997 TLGIVSTSPAAVRSAQRLAPLIEGT-TAREMSRVG---------------
MAV_1387|M.avium_104 GPFAAAMSQSVLLPAFIALFGIVAALFLVGFRPWAHRDGGTDAFGPDDYG
TH_3965|M.thermoresistible__bu APYAAAMAQSMLLPAFVALFGVLAAIFLAGIVLSTPQP---------DAD
Mb2361c|M.bovis_AF2122/97 DSANPRAHVGGIVPRRHIEHRDPVAIAEEDFIEG--------------IR
Rv2333c|M.tuberculosis_H37Rv DSANPRAHVGGIVPRRHIEHRDPVAIAEEDFIEG--------------IR
MMAR_1146|M.marinum_M VAHKVIDQLPAGPAAGVRDAVNHAFLDGLQVATLVCAG------------
MUL_0911|M.ulcerans_Agy99 VAHKVIDQLPAGPAAGVRDAVNHAFLDGLQVATLVCAG------------
MLBr_01562|M.leprae_Br4923 AGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL--------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1730|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5670|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0945|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_1387|M.avium_104 GDYHDDYDDDDAYVELILVREPEPEAQQRAGQPRRPQPTPAPPADVRRRD
TH_3965|M.thermoresistible__bu AQYLDEHDD---YLVIELAR-----------------PDPNPRTDIAVTA
Mb2361c|M.bovis_AF2122/97 VALLVATAT---LAVVFLAG-------------WRWFPRDVQTAGSDAKR
Rv2333c|M.tuberculosis_H37Rv VALLVATAT---LAVVFLAG-------------WRWFPRDVHTAGSDLSE
MMAR_1146|M.marinum_M ----------IALAAAIVVG----------------WLLPARAGQPTPAQ
MUL_0911|M.ulcerans_Agy99 ----------IALAAAIVVG----------------WLLPARAGQPTPAQ
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1730|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5670|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0945|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_1387|M.avium_104 PVESRRSVLDERPAQVQPIGFAHNGSHVDGGKRLRQVAVRRAPKPGPPAD
TH_3965|M.thermoresistible__bu VIPALRPTLRP--------------------------TLR--PSLGPPGG
Mb2361c|M.bovis_AF2122/97 --EAAYSDDRR--------------------------------VRG----
Rv2333c|M.tuberculosis_H37Rv RLPTAMTVECA--------------------------------VSHMPGA
MMAR_1146|M.marinum_M TEPALAQA------------------------------------------
MUL_0911|M.ulcerans_Agy99 TEPALAQV------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 -----------------------------------------------
Mflv_1730|M.gilvum_PYR-GCK -----------------------------------------------
MSMEG_5670|M.smegmatis_MC2_155 -----------------------------------------------
MAB_0945|M.abscessus_ATCC_1997 -----------------------------------------------
MAV_1387|M.avium_104 RFTRPPRRHHPGPAGHHLGEGESLRGQHHRPDPDDDPTGYGRHSSGN
TH_3965|M.thermoresistible__bu PVVQP-------------NNTAALR----------------------
Mb2361c|M.bovis_AF2122/97 -----------------------------------------------
Rv2333c|M.tuberculosis_H37Rv TWCRLWPA---------------------------------------
MMAR_1146|M.marinum_M -----------------------------------------------
MUL_0911|M.ulcerans_Agy99 -----------------------------------------------
MLBr_01562|M.leprae_Br4923 -----------------------------------------------