For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIQPVFRDHPIPALAVICLSVFVISVDATIVNVALPTLSRQLGADTAQLQWIVDAYTLVMSGLLLSAGSL SDRYGRRGWLSAGLILFALTSALAAQVNSADTLIAARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIG LWTAMVGVGVAVGPITGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLMLSAVG ITALVYTIIEAPNWGWTSTRASGGFIAAAIVLVGFALWERRSSHPMLDVSVFANRRFSGGSLAVTAGFLT LFGFIFVITQYFQFIKDYTALQTGVRLLPVALSIALAAILGPRLVERVGTTAVVAAGLAVFAAALAWAST ADATTPYTQIALQMLLLGGGLGLTTAPATEAIMGSLPADRAGVGSAVNDTTRELGGTLGVAIAGSVFASV YSGQLGSASTLATLPSDVRSAMKGSMAVAHKVIDQLPAGPAAGVRDAVNHAFLDGLQVATLVCAGIALAA AIVVGWLLPARAGQPTPAQTEPALAQA*
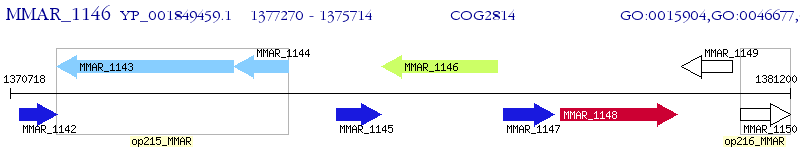
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1146 | - | - | 100% (518) | integral membrane transport protein |
| M. marinum M | MMAR_5094 | - | e-157 | 57.32% (492) | integral membrane transport protein |
| M. marinum M | MMAR_5192 | - | 3e-94 | 43.32% (434) | integral membrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2361c | - | 1e-73 | 33.94% (495) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_5055 | - | 3e-85 | 38.41% (492) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv2333c | - | 1e-72 | 33.74% (495) | integral membrane transport protein |
| M. leprae Br4923 | MLBr_01562 | - | 1e-46 | 29.37% (412) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_0945 | - | 2e-76 | 34.46% (444) | EmrB/QacA family drug resistance transporter |
| M. avium 104 | MAV_1387 | - | 6e-48 | 31.00% (400) | drug transporter |
| M. smegmatis MC2 155 | MSMEG_1427 | - | 9e-85 | 38.01% (492) | transmembrane efflux protein |
| M. thermoresistible (build 8) | TH_2682 | - | 4e-94 | 40.56% (498) | PUTATIVE transmembrane efflux protein |
| M. ulcerans Agy99 | MUL_0911 | - | 0.0 | 98.84% (517) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_0664 | - | 9e-87 | 37.10% (504) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1146|M.marinum_M --------------------------------------------MRIQPV
MUL_0911|M.ulcerans_Agy99 --------------------------------------------MRIQPV
MSMEG_1427|M.smegmatis_MC2_155 -------------------------------MTDALVEPGQNIDSRMAEL
TH_2682|M.thermoresistible__bu -------------------------------MVDALSQPGQDIGASVAAL
Mflv_5055|M.gilvum_PYR-GCK -------------------------------MVDTLPRSDQDIDAAVMTL
Mvan_0664|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2361c|M.bovis_AF2122/97 --------------------------------------------------
Rv2333c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0945|M.abscessus_ATCC_1997 ------------------------------------------MEAAVGAL
MAV_1387|M.avium_104 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
MMAR_1146|M.marinum_M FRDHPIPALAVICLSVFVISVDATIVNVALPTLSRQLGADTAQLQWIVDA
MUL_0911|M.ulcerans_Agy99 FRDHPILALAVICLSVFVISVDATIVNVALPTLSRELGADTAQLQWIVDA
MSMEG_1427|M.smegmatis_MC2_155 SVRQRYWLLIVACLDVSLVISSMVALNAALPDIATQTSATQAQLTWVIDG
TH_2682|M.thermoresistible__bu STRARVWLLTVACMGVALVISSMVALNAALPDIAVETAATQSQLTWVVDG
Mflv_5055|M.gilvum_PYR-GCK SKRRRIWLLAVASIDVLMVISSMVALNAALPDIALQTSATQTQLTWIVDG
Mvan_0664|M.vanbaalenii_PYR-1 -MTRRWSALAVLTLAVLLIGIDGTVLALATPFISADLGATGTQILWIGDI
Mb2361c|M.bovis_AF2122/97 MNRTQLLTLIATGLGLFMIFLDALIVNVALPDIQRSFAVGEDGLQWVVAS
Rv2333c|M.tuberculosis_H37Rv MNRTQLLTLIATGLGLFMIFLDALIVNVALPDIQRSFAVGEDGLQWVVAS
MAB_0945|M.abscessus_ATCC_1997 SERRKIVILGSCCLSLLIVSMDSTIVNVALPTIRADFGATTSQLQWVIDI
MAV_1387|M.avium_104 -----------MMIGFFMIMVDSTIVAIANPTIMADLHIGYDTVVWVTSA
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
: . * * : . *
MMAR_1146|M.marinum_M YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLIA
MUL_0911|M.ulcerans_Agy99 YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLVA
MSMEG_1427|M.smegmatis_MC2_155 YTLVLACLLLPAGAVGDRYGRRGALLIGLAIFSLASAAPLIFDSPVQLIV
TH_2682|M.thermoresistible__bu YTLVLACLLLPAGAIGDRYGRRGAMLVGLGLFTVASAAPLVLDGPLTLIL
Mflv_5055|M.gilvum_PYR-GCK YTLALACLLLPAGALGDRYGRRGALLVGLAIFALASLAPVFLDSPTQIIV
Mvan_0664|M.vanbaalenii_PYR-1 YSFVLAALLISMGSLGDRIGHRKLLLCGATAFAVISAVTAYSTSPQMLIL
Mb2361c|M.bovis_AF2122/97 YSLGMAVFIMSAATLADLYGRRRWYLIGVSLFTLGSIACGLAPSIAVLTT
Rv2333c|M.tuberculosis_H37Rv YSLGMAVFIMSAATLADLDGRRRWYLIGVSLFTLGSIACGLAPSIAVLTT
MAB_0945|M.abscessus_ATCC_1997 YTLGLASLLMLSGAAGDRLGRRKVFHLGLSIFAIGSLLCSVAPTVGLLIA
MAV_1387|M.avium_104 YLLGYAVVLLVAGRLGDRFGTKNLYLIGLAVFTVASVWCGLAGSAAMLIA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
* : . .:: . .* * : * *:: * :
MMAR_1146|M.marinum_M ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
MUL_0911|M.ulcerans_Agy99 ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
MSMEG_1427|M.smegmatis_MC2_155 ARAVAGAGAAFIMPATLSLLTAAFPK-SERNKAVGVWAGVVGSGAIVGFL
TH_2682|M.thermoresistible__bu SRMVAGVGAAFVMPATLSLLTAAYPR-AQRNKAVGIWAAVAGSGAVVGFL
Mflv_5055|M.gilvum_PYR-GCK ARAVAGVGAALIMPATLSLLTSAFPK-SERNKAVGIWAGVAGSGAVFGFL
Mvan_0664|M.vanbaalenii_PYR-1 TRALLGMAGATLTPATLALIRGLFPDQRERSIAVGIWASAFSAGAALGPV
Mb2361c|M.bovis_AF2122/97 ARGAQGLGAAAVSVTSLALVSAAFPEAKEKARAIGIWTAIASIGTTTGPT
Rv2333c|M.tuberculosis_H37Rv ARGAQGLGAAAVSVTSLALVSAAFPEAKEKARAIGIWTAIASIGTTTGPT
MAB_0945|M.abscessus_ATCC_1997 ARLLQAVGGSMLNPVALSIISQVFTGRVERARALGFWGAVVGISMALGPI
MAV_1387|M.avium_104 ARVVQGVGAGVMTPQTLSTITRIFPA-ERRGVAVSVWSATAGAASLAGPL
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
:* . ... *. : :. : * ..: . . . *
MMAR_1146|M.marinum_M TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLM
MUL_0911|M.ulcerans_Agy99 TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLM
MSMEG_1427|M.smegmatis_MC2_155 VTGGLLHFWTWQSIFVT-FTIASVGLFIVTCTVPSSRDDSAAPLDWLGAA
TH_2682|M.thermoresistible__bu GTGVLLHFWSWPSIFWA-FTVSGAGLFVLACTVASSRDHRATPLDWPGAV
Mflv_5055|M.gilvum_PYR-GCK GTGVLLHFFSWQSIFYM-FAAGALLMFIATCTIGSSRDETATPIDWVGGA
Mvan_0664|M.vanbaalenii_PYR-1 VGGVLLEHFWWGSVFLINIPVMVILLVGGVLLLPEHRNPDPGPWDLPSVA
Mb2361c|M.bovis_AF2122/97 LGGLLVDQWGWRSIFYVNLPMGALVLFLTLCYVEESCNERARRFDLSGQL
Rv2333c|M.tuberculosis_H37Rv LGGLLVDQWGWRSIFYVNLPMGALVLFLTLCYVEESCNERARRFDLSGQL
MAB_0945|M.abscessus_ATCC_1997 VGGLLIQWVGWRAVFWINLPVCVLAFVLTAVFVPETKSATMRDIDPVGQA
MAV_1387|M.avium_104 AGGVLVDGLGWQWIFFVNVPIGVLGLVLAYWLVPVLPTQSHR-FDLVGVG
MLBr_01562|M.leprae_Br4923 VGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKERMK-LDAAGAI
* * . * * .. : : * .
MMAR_1146|M.marinum_M LSAVGITALVYTIIEAPN--WGWTSTRASGGFIAAAIVLVGFALWE-RRS
MUL_0911|M.ulcerans_Agy99 LSAVGITALVYTIIEAPN--WGWTSTRASGGFIAAAIVLVGFALWE-RRS
MSMEG_1427|M.smegmatis_MC2_155 LIGTAVAAFVFGVVEAPV--RGWTHPSVWGAMAGGVALAAAFTAVQ-LRR
TH_2682|M.thermoresistible__bu LIGGAVAVFVYGVLEAPV--HGWTHPVVYGSMAAGVALALAFAVVE-LRS
Mflv_5055|M.gilvum_PYR-GCK LIGTAIAVFVLGVVEAPA--RGWTDPVVLGCLAAGVVLAVAFAMVQ-LRR
Mvan_0664|M.vanbaalenii_PYR-1 LSMVGMLAVVYAVKEGFAGLAHGVGPDVAVAGLVGAAALTMFVRRQ-LRL
Mb2361c|M.bovis_AF2122/97 LFIVAVGALVYAVIEGPQ--IGWTSVQTIVMLWTAAVGCALFVWLE-RRS
Rv2333c|M.tuberculosis_H37Rv LFIVAVGALVYAVIEGPQ--IGWTSVQTIVMLWTAAVGCALFVWLE-RRS
MAB_0945|M.abscessus_ATCC_1997 LAVLFLFGIVFALIEGPS--LGWATPGLIVVLAVALAAFAVFLRYE-SRR
MAV_1387|M.avium_104 LSGVGMLLIVFGLQQGQA---AHWQPWIWALIVAGVGFVTVFVFWQSVNV
MLBr_01562|M.leprae_Br4923 LATLACTAAVFAFSMGPE--KGWISLTTIGSGLVALVAAIAFAVVE-RTA
* * . . . * :
MMAR_1146|M.marinum_M SHPMLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQ
MUL_0911|M.ulcerans_Agy99 SHPMLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQ
MSMEG_1427|M.smegmatis_MC2_155 THPLLDVRLFRDPNFATGSAGVTFLFFATFGFFFVSMQYIQLIMGYSPIA
TH_2682|M.thermoresistible__bu AHPLLDIRLFAKPDFTTGAVGVTFFFLANFGFFFVSMQYMQLMLGYSALQ
Mflv_5055|M.gilvum_PYR-GCK EHPLLDVRLFTRPDFATGAVGITFLFMANFGFFFIEMQFMQLVMGYSALE
Mvan_0664|M.vanbaalenii_PYR-1 PAPLIDVRLFANPRFSGVVAANLLSVLGLSGLVFFLSQYFQLVHGYGPLK
Mb2361c|M.bovis_AF2122/97 SNPMMDLTLFRDTSYALAIATICTVFFAVYGMLLLTTQFLQNVRGYTPSV
Rv2333c|M.tuberculosis_H37Rv SNPMMDLTLFRDTSYALAIATICTVFFAVYGMLLLTTQFLQNVRGYTPSV
MAB_0945|M.abscessus_ATCC_1997 RHPFIDLRFFRSIPFASATLTAICACMAWSAFLFTMSLYLQGQRHFSAMH
MAV_1387|M.avium_104 REPLIPLVIFSDRDFSLCNIGVAIISFAATAMMLPLTFYAQAVCGLSPTR
MLBr_01562|M.leprae_Br4923 ENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALR
*.: . .* .: . : * .
MMAR_1146|M.marinum_M TGVRLLPVALSIALAAILGPRLVERVGTTAVVAAGLAVFAAALAWAST--
MUL_0911|M.ulcerans_Agy99 TGVRLLPVAFSIALAAILGPRLVERVGTTAVVAAGLAVFAAALAWAST--
MSMEG_1427|M.smegmatis_MC2_155 TAIALCPLVVPILAFGATTHLYLPLLGLRLTAFLGLLVTAVGLWCMCT--
TH_2682|M.thermoresistible__bu TAFALTPLTIPLLLLSVTTQWYLPRLGLRAVISTGLLLIAAGLFYART--
Mflv_5055|M.gilvum_PYR-GCK TAFALAPLAFPVLVLGGTLHLYLPKVGLRFAVTGGLLLLAGGLFLMRF--
Mvan_0664|M.vanbaalenii_PYR-1 AGLAELPAAVTATVFGVLAGVAVRYVSHRAVLATGLALVGVAMAAMTTSL
Mb2361c|M.bovis_AF2122/97 TGLMILPFSAAVAIVSPLVGHLVGRIGARVPILAG-LCMLMLGLLMLI--
Rv2333c|M.tuberculosis_H37Rv TGLMILPFSAAVAIVSPLVGHLVGRIGARVPILAG-LCMLMLGLLMLI--
MAB_0945|M.abscessus_ATCC_1997 TGLIYLPIAVGALVFSPISGRMVGRFGARPSLLAAGTLFLTASLLLTR--
MAV_1387|M.avium_104 SALLIAPMAIANGVFAPFVGKIVDRYHPRPVLGFGFSLLAIALTWLTF--
MLBr_01562|M.leprae_Br4923 AGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWA--
:.. * : .
MMAR_1146|M.marinum_M --ADATTPYTQ-IALQMLLLGGGLGLTTAPATEAIMGSLPADRAGVGSAV
MUL_0911|M.ulcerans_Agy99 --ADATTPYTQ-ITLQMLLLGGGLGLTTAPATEAIMGSLPADRAGVGSAV
MSMEG_1427|M.smegmatis_MC2_155 --LETGSTYFD-FAWPLFIMSIGVGLSTAPVTSAIMGAAPDEKQGVASAV
TH_2682|M.thermoresistible__bu --LEIASPYLD-LAVPLLIMSTGIGLCTAPSTSAIMGAVPDEKHGVASAV
Mflv_5055|M.gilvum_PYR-GCK --LDADATFLD-IMWPLLLAASGIGLCTAPTTSAIMNAAPDEKQGVASAV
Mvan_0664|M.vanbaalenii_PYR-1 LVFTPVAPYLP-LGVALFVVGVGLGLAFTVASDVILASVPRERAGAAAAV
Mb2361c|M.bovis_AF2122/97 --FSEHRSSAL-VLVGLGLCGSGVALCLTPITTVAMTAVPAERAGMASGI
Rv2333c|M.tuberculosis_H37Rv --FSEHRSSAL-VLVGLGLCGSGVALCLTPITTVAMTAVPAERAGMASGI
MAB_0945|M.abscessus_ATCC_1997 --VSEATPIWE-LLVTFAIFGAGFAMVNAPITNAAVSGMPLDRAGAASAV
MAV_1387|M.avium_104 -EMSPATPIWR-LVLPFFAMGVGMAFVWSPLTATATRNLSAQLAGAGSAV
MLBr_01562|M.leprae_Br4923 -FMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAI
. . : . *..: : : * :.:
MMAR_1146|M.marinum_M NDTTRELGGTLGVAIAGSVFASVYSGQLGS-------ASTLATLPSDVRS
MUL_0911|M.ulcerans_Agy99 NDTTRELGGTLGVAIAGSVFASVYSGQLGS-------ASALATLPSDVRS
MSMEG_1427|M.smegmatis_MC2_155 NDTTREIGAALGIAVAGSVLAAQYERHL---------SPALGAMPEAVRG
TH_2682|M.thermoresistible__bu NDTTREIGAALGIAVAGSVLAAEYGDRL---------RPHLGALPEPVRQ
Mflv_5055|M.gilvum_PYR-GCK NDATREIGAALGIAVAGSILAAVYHRSL---------ASRLADFPEQIRT
Mvan_0664|M.vanbaalenii_PYR-1 SETAYELGMALGIAMLGSIVTAVYRTVV-----------IAPEIPSAAAD
Mb2361c|M.bovis_AF2122/97 MSAQRAIGSTIGFAVLGSVLAAWLSATLEP-------HLERAVPDPVQRH
Rv2333c|M.tuberculosis_H37Rv MSAQRAIGSTIGFAVLGSVLAAWLSATLEP-------HLERAVPDPVQRH
MAB_0945|M.abscessus_ATCC_1997 TSTSRQVGVSIGVALCGSIGTPALWFMCAG--------------------
MAV_1387|M.avium_104 YNSVRQLGAVLGSAGMAAFMTWRIGAEMPGQPGGGGEDSAGPVLPEFLRG
MLBr_01562|M.leprae_Br4923 ALMLQSLGGPLVLAVIQAVITSRTLYLG----------------------
:* : * :. :
MMAR_1146|M.marinum_M AMKGSMAVAHKVIDQLPAG-------------------------------
MUL_0911|M.ulcerans_Agy99 AMKGSMAVAHKVIDQLPAG-------------------------------
MSMEG_1427|M.smegmatis_MC2_155 PALNSLAEALTAAEHMGP--------------------------------
TH_2682|M.thermoresistible__bu AASNSLAEALEAARQLGP--------------------------------
Mflv_5055|M.gilvum_PYR-GCK TATDSLAHALSISEQMGP--------------------------------
Mvan_0664|M.vanbaalenii_PYR-1 LARDSLPGAIHAAHSLPAD-------------------------------
Mb2361c|M.bovis_AF2122/97 VLAEIIIDSANPRAHVGGI-------------------------------
Rv2333c|M.tuberculosis_H37Rv VLAEIIIDSANPRAHVGGI-------------------------------
MAB_0945|M.abscessus_ATCC_1997 ---------------LGLL-------------------------------
MAV_1387|M.avium_104 PFAAAMSQSVLLPAFIALFGIVAALFLVGFRPWAHRDGGTDAFGPDDYGG
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MMAR_1146|M.marinum_M -------------------------------PAAGVRDAVNHAFLDGLQV
MUL_0911|M.ulcerans_Agy99 -------------------------------PAAGVRDAVNHAFLDGLQV
MSMEG_1427|M.smegmatis_MC2_155 -------------------------------QGAELAQTARQAFVDATGS
TH_2682|M.thermoresistible__bu -------------------------------QGARLAEFAQSAFLEAMHV
Mflv_5055|M.gilvum_PYR-GCK -------------------------------QGARLSELAQDAFIHAADR
Mvan_0664|M.vanbaalenii_PYR-1 -------------------------------QQSTLLDAARASFTHGLAS
Mb2361c|M.bovis_AF2122/97 -------------------------------VPRRHIEHRDPVAIAEEDF
Rv2333c|M.tuberculosis_H37Rv -------------------------------VPRRHIEHRDPVAIAEEDF
MAB_0945|M.abscessus_ATCC_1997 -------------------------------IATLGIVSTSPAAVR----
MAV_1387|M.avium_104 DYHDDYDDDDAYVELILVREPEPEAQQRAGQPRRPQPTPAPPADVRRRDP
MLBr_01562|M.leprae_Br4923 -------------------------------GTTGPVKFMNDAQLRALDH
MMAR_1146|M.marinum_M ATLVCAGIALAAAIVVGWLL-PARAGQPTPAQTEPALAQA----------
MUL_0911|M.ulcerans_Agy99 ATLVCAGIALAAAIVVGWLL-PARAGQPTPAQTEPALAQV----------
MSMEG_1427|M.smegmatis_MC2_155 AVIILGSVVAVAAVLTGIWG-PGRDGQQLRVIRRLRSR------------
TH_2682|M.thermoresistible__bu SLIVLGALCALAAAFVAVWA-PGRDGRQYGFVERWTSRWSGEEAGGAVAE
Mflv_5055|M.gilvum_PYR-GCK ALLVLSLLLLVAAVFVAIWS-PGRDGRQWSVVRRLRKN------------
Mvan_0664|M.vanbaalenii_PYR-1 ASAAGAVLLLTAAVAVWWLLRPGSPGAAEVGVLADPGVHRDPGRDARVDA
Mb2361c|M.bovis_AF2122/97 IEGIRVALLVATATLAVVFLAGWRWFPRDVQTAGSDAKR--EAAYSDDRR
Rv2333c|M.tuberculosis_H37Rv IEGIRVALLVATATLAVVFLAGWRWFPRDVHTAGSDLSERLPTAMTVECA
MAB_0945|M.abscessus_ATCC_1997 -SAQRLAPLIEGTTAREMSRVG----------------------------
MAV_1387|M.avium_104 VESRRSVLDERPAQVQPIGFAHNGSHVDGGKRLRQVAVRRAPKPGPPADR
MLBr_01562|M.leprae_Br4923 GYTYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL-----
.
MMAR_1146|M.marinum_M ----------------------------------------------
MUL_0911|M.ulcerans_Agy99 ----------------------------------------------
MSMEG_1427|M.smegmatis_MC2_155 ----------------------------------------------
TH_2682|M.thermoresistible__bu HDDGGVRAATGDGWEHGTVDDP------------------------
Mflv_5055|M.gilvum_PYR-GCK ----------------------------------------------
Mvan_0664|M.vanbaalenii_PYR-1 PGGAELGDRNS-----------------------------------
Mb2361c|M.bovis_AF2122/97 VRG-------------------------------------------
Rv2333c|M.tuberculosis_H37Rv VSHMPGATWCRLWPA-------------------------------
MAB_0945|M.abscessus_ATCC_1997 ----------------------------------------------
MAV_1387|M.avium_104 FTRPPRRHHPGPAGHHLGEGESLRGQHHRPDPDDDPTGYGRHSSGN
MLBr_01562|M.leprae_Br4923 ----------------------------------------------