For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQLFVTGGSGFVGQHLIRRLVGAGHEVRALARTDSAAGLVGRVGAEPVLGDLSDLVNSDPPPLWASALRG VDAVVHGAAYMAFWGPDDVFRRANLEPSVALHQVAASSGVTRFVLISAASVATGTQRAPVVDERTDEGRP NIAYSRVKLATERILLNAVTPTMTTVALRPPFIWGAGMSTLADFVAAVEAGRFSWIDNGKHTVDFVHVDN LAKAVGLALTRGRAGRAYYITDGAPMPTRDFFTPLLATQGVDVSAARSVPFALAAPLGALLEAGARLLRR PEPPMLTNWITTFMGRDRSYDITAARSELDYTPSVNLADGLAEMADSLSR
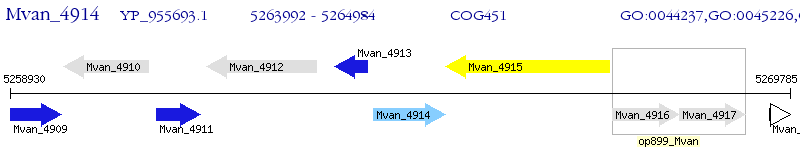
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4914 | - | - | 100% (330) | NAD-dependent epimerase/dehydratase |
| M. vanbaalenii PYR-1 | Mvan_4635 | - | 8e-19 | 32.71% (266) | 3-beta hydroxysteroid dehydrogenase/isomerase |
| M. vanbaalenii PYR-1 | Mvan_4271 | - | 1e-14 | 28.12% (320) | NAD-dependent epimerase/dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1136c | - | 4e-19 | 29.24% (342) | cholesterol dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2075 | - | 1e-21 | 31.58% (342) | 3-beta hydroxysteroid dehydrogenase/isomerase |
| M. tuberculosis H37Rv | Rv1106c | - | 4e-19 | 29.24% (342) | cholesterol dehydrogenase |
| M. leprae Br4923 | MLBr_01942 | - | 1e-19 | 28.78% (344) | putative cholesterol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0718c | - | 3e-14 | 25.39% (319) | NAD-dependent epimerase/dehydratase |
| M. marinum M | MMAR_3324 | - | 9e-20 | 26.83% (328) | nucleoside-diphosphate-sugar epimerase |
| M. avium 104 | MAV_1225 | - | 3e-20 | 30.41% (342) | 3-beta hydroxysteroid dehydrogenase/isomerase family protein |
| M. smegmatis MC2 155 | MSMEG_5228 | - | 1e-21 | 31.61% (329) | 3-beta hydroxysteroid dehydrogenase/isomerase family protein |
| M. thermoresistible (build 8) | TH_1114 | - | 2e-22 | 31.70% (347) | PROBABLE CHOLESTEROL DEHYDROGENASE |
| M. ulcerans Agy99 | MUL_0172 | - | 4e-20 | 28.45% (341) | cholesterol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1136c|M.bovis_AF2122/97 -MLRRMGDASLTTELGRVLVTGGAGFVGANLVTTLLDRGHWVRSFDR--A
Rv1106c|M.tuberculosis_H37Rv -MLRRMGDASLTTELGRVLVTGGAGFVGANLVTTLLDRGHWVRSFDR--A
MAV_1225|M.avium_104 -MIGPMGDPSLTAELGRVLVTGGSGFVGANLVTTLLERGYQVRSFDR--A
MUL_0172|M.ulcerans_Agy99 -MLRPMGDASLTTELGRVLVTGGSGFVGSNLVTTLLDRGHHVRSFDR--A
MLBr_01942|M.leprae_Br4923 -MLRGVGNASLTTELGRVLVTGGSGFVGANLVTALLERGYQVRSFDR--A
TH_1114|M.thermoresistible__bu -----MGDPTLRTDLGRVLVTGGSGFVGTNLVTTLLERGHHVRSCDR--V
Mflv_2075|M.gilvum_PYR-GCK -----MLR----TELGRVLVTGGSGFVGANLVTELLDRGLHVRSFDR--V
MSMEG_5228|M.smegmatis_MC2_155 -----MADS--TTDLGRILVTGGSGFVGANLVTELLDRGYAVRSFDR--V
Mvan_4914|M.vanbaalenii_PYR-1 ---------------MQLFVTGGSGFVGQHLIRRLVGAGHEVRALARTDS
MMAR_3324|M.marinum_M MSTDQSARPAPFFAQRKIFITGANGFIGANLAARLRQLGARVTGVDL---
MAB_0718c|M.abscessus_ATCC_199 ---------------MKTAVTGAAGFIGTNLVNLLVDNGHEVIAIDR-VV
: :**. **:* :* * * * .
Mb1136c|M.bovis_AF2122/97 PSLLPAHPQLEVLQGDITDAD------VCAAAVDGIDTIFHTAAIIELMG
Rv1106c|M.tuberculosis_H37Rv PSLLPAHPQLEVLQGDITDAD------VCAAAVDGIDTIFHTAAIIELMG
MAV_1225|M.avium_104 PSPLPAHPHLEVLQGDITDAG------VCAAAVEGIDTIFHTAAIIDLMG
MUL_0172|M.ulcerans_Agy99 PSPLPPHPQLEVLQGDITDAA------TCATAVDGVDTVFHTAAIIELMG
MLBr_01942|M.leprae_Br4923 PMPLPQHPQLEVLQGDITDAT------VCTTAMDSIDTVFHTAAIIELMG
TH_1114|M.thermoresistible__bu GSPLPDHPRLEVIDGDICDAD------AVAAAVDGIDTVFHTAAVIDLMG
Mflv_2075|M.gilvum_PYR-GCK ASALPAHARLEIVEGDITDAD------DVAAAVEGVDTVFHTAAIIDLMG
MSMEG_5228|M.smegmatis_MC2_155 PSPLPAHAGLEVATGDICDLD------NVTNAVAGVDTVFHTAAIIDLMG
Mvan_4914|M.vanbaalenii_PYR-1 AAGLVGRVGAEPVLGDLSDLVNSDPPPLWASALRGVDAVVHGAAYMAFWG
MMAR_3324|M.marinum_M ----VADPANGIIAGSTADPA------AWASALDGVDAVVHLAALV----
MAB_0718c|M.abscessus_ATCC_199 PASPETREAVTWVSGDVLDPD------SMTKALDGVEVVYHLVAVIT---
*. * : *: .::.: * .* :
Mb1136c|M.bovis_AF2122/97 GASVTDEYRQRSFAVNVGGTENLLHAGQRAGVQRFVYTSSNSVVMGGQNI
Rv1106c|M.tuberculosis_H37Rv GASVTDEYRQRSFAVNVGGTENLLHAGQRAGVQRFVYTSSNSVVMGGQNI
MAV_1225|M.avium_104 GASVTEEYRQRSFAVNVGGTENLVRAGQAAGVQRFVYTSSNSVVMGGQNI
MUL_0172|M.ulcerans_Agy99 GASVTDEYRKRSFSVNVGGTENLVRAGQQAGVKRFVYTSSNSVVMGGQII
MLBr_01942|M.leprae_Br4923 GASVTDEYRQRSYTVNVGGTENLLRAGQKSGVKRFVYTASNSVVMGGTPI
TH_1114|M.thermoresistible__bu GAGVTDEYRERSFAVNVHGTERLVRAAQQAGVRRFVYTASNSVVMGGQPI
Mflv_2075|M.gilvum_PYR-GCK GASVSEEYRQRSFAVNVTGTQNLVHAAQKAGVTRFVYTASNSVVMGGQRI
MSMEG_5228|M.smegmatis_MC2_155 GASVTAEYRQRSFAVNVGGTENLVRAAQSAGVKRFVYTASNSVVMGGQHI
Mvan_4914|M.vanbaalenii_PYR-1 ---PDDVFRR----ANLEPSVALHQVAASSGVTRFVLISAASVATGTQRA
MMAR_3324|M.marinum_M ---STVVAVETAWDVNVLGTKKVIDAAVDAGVRRFVHLSSIAAYGWDFPD
MAB_0718c|M.abscessus_ATCC_199 ----LKHEDELCWRINTEGARTVAQAALTVGARRMVHCSSIDSYSN-SVA
. * : : .. *. *:* ::
Mb1136c|M.bovis_AF2122/97 AGGDETLPYTDRFNDLYTETKVVAERFVLAQNGV---DGMLTCAIRPSGI
Rv1106c|M.tuberculosis_H37Rv AGGDETLPYTDRFNDLYTETKVVAERFVLAQNGV---DGMLTCAIRPSGI
MAV_1225|M.avium_104 VGGDETLPYTDRFNDLYTETKVLAERFVLGQNGV---DGMLTCAIRPSGI
MUL_0172|M.ulcerans_Agy99 AGGDETLPYTNRFNDLYTETKVVAEKFVLSQNGN---NEMLTCAIRPSGI
MLBr_01942|M.leprae_Br4923 TGGDETMPYTKRFNDLYTETKVVAEKFVLSQNGVPDGETMLTCSIRPSGI
TH_1114|M.thermoresistible__bu ADGDETLPYTDRFNDLYTETKVIAERFVLGQNGI---EGMLTCSIRPSGI
Mflv_2075|M.gilvum_PYR-GCK AGGDETLPYTERFNDLYTETKVVAEKFVLSQNGV---SGLLTCSIRPSGI
MSMEG_5228|M.smegmatis_MC2_155 VHGDETLPYTERFNDLYTETKVVAEKFVLGRNGV---AGMLTCSIRPSGI
Mvan_4914|M.vanbaalenii_PYR-1 PVVDERTD-EGRPNIAYSRVKLATERILLNAVTP----TMTTVALRPPFI
MMAR_3324|M.marinum_M HVTEDYPTRVTGGLSTYVDTKTNSELVALANANR----GMEMVVVRPADV
MAB_0718c|M.abscessus_ATCC_199 TIDEKSPRSAAADLPVYQRSKWGGEVAVRETIAS----GLDAVICNPTGV
:. * * * : .*. :
Mb1136c|M.bovis_AF2122/97 WGNGDQTMFRKLFESVLKGHVKVLVGRKSARLDNSYVHNLIHGFILAAAH
Rv1106c|M.tuberculosis_H37Rv WGNGDQTMFRKLFESVLKGHVKVLVGRKSARLDNSYVHNLIHGFILAAAH
MAV_1225|M.avium_104 WGRGDQTMFRKLFESVIAGHVKVLIGRKSARLDNSYVHNLIHGFILAAQH
MUL_0172|M.ulcerans_Agy99 WGTGDQLMFRKLFESVIKGHVKVLVGPKSVLLDNSYVDNLIHGFMLAAQH
MLBr_01942|M.leprae_Br4923 WGRGDQTMFRKAFESVVSGHVKVLIGSKNAKLDNSYVHNLVHGLILAAEH
TH_1114|M.thermoresistible__bu WGPGDQTMFRKLFESVHAGHVKVLIGSRHARLDNSYVHNLVHGFILAAEH
Mflv_2075|M.gilvum_PYR-GCK WGRGDQTMFRKVFESVLAGHVKVLVGNKETKLDNSYVHNLVHGFILAAEH
MSMEG_5228|M.smegmatis_MC2_155 WGRGDQTMFRKVFESVLAGHVKVLVGRKSTLLDNSYVHNLVHGFILAAEH
Mvan_4914|M.vanbaalenii_PYR-1 WGAG-MSTLADFVAAVEAGRFSWIDNGK-HTVDFVHVDNLAK-----AVG
MMAR_3324|M.marinum_M YGPG--SVWIREPIAMAKANQLILPERGSGVFDVIYIDNFVD----AMVL
MAB_0718c|M.abscessus_ATCC_199 YGPVDLPNLSRINQLLFDSARGRLPGMVNSQYDLVDARDVAAG-----LY
:* : . : * :.
Mb1136c|M.bovis_AF2122/97 LVPDGTAPGQAYFINDAEPINMFEFARPVLEACGQRWPKMRISGPAVRWV
Rv1106c|M.tuberculosis_H37Rv LVPDGTAPGQAYFINDAEPINMFEFARPVLEACGQRWPKMRISGPAVRWV
MAV_1225|M.avium_104 LTPGGTAPGQAYFINDAEPINMFEFARPVVEACGVNWPRVRVNGPIVRAA
MUL_0172|M.ulcerans_Agy99 LVPGGSAPGQAYFINDAEPINMFDFARPVVEACGEKWPRVRVSGPMVHRA
MLBr_01942|M.leprae_Br4923 LVPGGTAPGQAYFINDGEPINFFDFMGPIIKACGENWPRVRISGRLVRNV
TH_1114|M.thermoresistible__bu LVPGGTAPGQAYFINDGEPINMFEFARPVVTACGQRWPTLRVSGPLVRAV
Mflv_2075|M.gilvum_PYR-GCK LVEGGSAPGQAYFINDGEPINMFEFARPVVQACGEPFPKFRVPGRLVWFA
MSMEG_5228|M.smegmatis_MC2_155 LTPNGTAPGQAYFINDGEPVNMFEFARPVIEACGRKLPRVRVPGRAVHAA
Mvan_4914|M.vanbaalenii_PYR-1 LALTRGRAGRAYYITDGAPMPTRDFFTPLLATQGVDVSAARSVPFALAAP
MMAR_3324|M.marinum_M VLATEGIAGEVFNLGEELAVSCQEYFGEVASWTGAKVRSVPIR--IGAPA
MAB_0718c|M.abscessus_ATCC_199 LAGEKGRTGENYLLG-GHMGSLLQVCRLAARRAGKRGPRFAVPMGLLNAA
: .*. : : : *
Mb1136c|M.bovis_AF2122/97 MTGWQRLHFRFG--FPAPLLEPLAVERLYLDNYFSIAKARRDLGYEPLFT
Rv1106c|M.tuberculosis_H37Rv MTGWQRLHFRFG--FPAPLLEPLAVERLYLDNYFSIAKARRDLGYEPLFT
MAV_1225|M.avium_104 MTGWQRLHFRFG--IPAPLLEPLAVERLYLDNFFSIAKASRDLGYQPLFT
MUL_0172|M.ulcerans_Agy99 MTGWQRLHFRFG--LPAPLLEPLAVERLYLDNYFSVDKARRDLGYEPKFT
MLBr_01942|M.leprae_Br4923 MAVWQRLHFGFG--LPKPPMEPLAVERVYLDNYFSIEKAHKELGYRPLFT
TH_1114|M.thermoresistible__bu MSLWQRLHFRFG--LPKPPLEPLAVERLYLDNYFSIDKARRELGYRPRYT
Mflv_2075|M.gilvum_PYR-GCK MTIWQFLHFKFG--LPKPLLEPLAVERLYLDNYFSIAKAQRDLGYQPLFT
MSMEG_5228|M.smegmatis_MC2_155 MSGWQRLHFRFG--IPEPLLEPLAVERLYLNNYFSIAKATRDLGYRPLFT
Mvan_4914|M.vanbaalenii_PYR-1 LGALLEAGARLLRRPEPPMLTNWITTFMGRDRSYDITAARSELDYTPSVN
MMAR_3324|M.marinum_M LGAIGRIQRRLG---MSSELGPALLHMLNRRYVVSNDKARDRLGFKPVVS
MAB_0718c|M.abscessus_ATCC_199 IPVIEPIGKLLK----NDALSRAAFNKLTVSPAVDITKARTELGYEPR-S
: : : : . * *.: * .
Mb1136c|M.bovis_AF2122/97 TQQALTECLPYYVSLFEQMKNEARAEKTAATVKP----
Rv1106c|M.tuberculosis_H37Rv TQQALTECLPYYVSLFEQMKNEARAEKTAATVKP----
MAV_1225|M.avium_104 TEQAMSECLPYYVGMFEQMKRQALAGKASA--------
MUL_0172|M.ulcerans_Agy99 TEQALKECLPYYVDLFRQMKSEVKAGGR----------
MLBr_01942|M.leprae_Br4923 TEQAMAECLPYYTELFEQIKVAAKPHLASIAATPRPE-
TH_1114|M.thermoresistible__bu TEQALQECLPYYVDLFHRMKAEPATV------------
Mflv_2075|M.gilvum_PYR-GCK TEQALAQCIPYYVELFDRMKREAGNPVVQAAAPAPPKG
MSMEG_5228|M.smegmatis_MC2_155 TEQARVDCLPYYVDLFKQMEAQAR--------PA----
Mvan_4914|M.vanbaalenii_PYR-1 LADGLAEMADSLSR------------------------
MMAR_3324|M.marinum_M YHEGMARSKEWARHEGLI--------------------
MAB_0718c|M.abscessus_ATCC_199 TETTVNEFVDFLLGSGRL--------------------