For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRVVGLGYLIIEATDLEEWQTFACDLLGLQAVVRTPDRLSLRVDEKAYRIDVRQAESDAVAVIGWEVRGP RELGQLANLLQANGYDVKWAGAEVARERRVSGLAGFTDPDGQAVELFWGLQEHRDRFVSPTGARFVTGAA GLGHVFQAVDDADVYSELYQNLLGFKLSDIIDFGPGASATFLHTTERHHSYAFAALPGLPNGVGHVMLEV DDLDIVGRAWEKVRDGAAPMVLDFGKHTNDEMLSFYVKTPSGFELEYGYGGKTIDDATWTPSRYDAASYW GHRRTRPTDPDSAELSV*
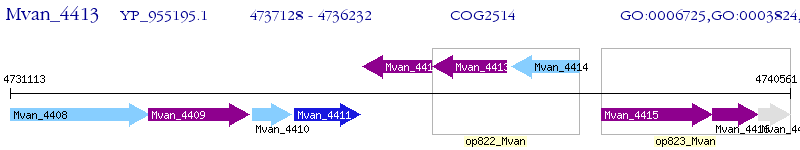
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4413 | - | - | 100% (298) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. vanbaalenii PYR-1 | Mvan_2891 | - | 2e-50 | 36.64% (292) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. vanbaalenii PYR-1 | Mvan_0545 | - | 3e-50 | 36.52% (293) | glyoxalase/bleomycin resistance protein/dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3599c | bphC | 2e-48 | 37.50% (288) | putative biphenyl-2,3-diol 1,2-dioxygenase BPHC (23OHBP |
| M. gilvum PYR-GCK | Mflv_0538 | - | 6e-50 | 36.55% (290) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | Rv3568c | bphC | 2e-48 | 37.50% (288) | biphenyl-2,3-diol 1,2-dioxygenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0076 | - | 8e-52 | 39.93% (288) | putative 2,3-dihydroxybiphenyl 1,2-dioxygenase or |
| M. marinum M | MMAR_5063 | bphC | 9e-48 | 38.19% (288) | biphenyl-2,3-diol 1,2-dioxygenase BphC |
| M. avium 104 | MAV_0593 | - | 5e-49 | 38.19% (288) | extradiol ring-cleavage dioxygenase |
| M. smegmatis MC2 155 | MSMEG_2891 | - | 2e-49 | 38.06% (289) | biphenyl-2,3-diol 1,2-dioxygenase 1 |
| M. thermoresistible (build 8) | TH_4499 | - | 5e-52 | 38.70% (292) | PUTATIVE biphenyl-2,3-diol 1,2-dioxygenase 1 |
| M. ulcerans Agy99 | MUL_4139 | bphC | 6e-48 | 38.19% (288) | biphenyl-2,3-diol 1,2-dioxygenase BphC |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5063|M.marinum_M -----MS---IRSLGYLRIEATDMAAWREYGLKVLGMVEGKGSTEGALYL
MUL_4139|M.ulcerans_Agy99 -----MS---IRSLGYLRIEATDMAAWREYGLKVLGMVEGKGSTEGALYL
MAV_0593|M.avium_104 -----MSNGAIRSLGYLRIEATDMAAWRDYGLKVLGMVEGKGDTEGALYL
Mb3599c|M.bovis_AF2122/97 -----MS---IRSLGYLRIEATDMAAWREYGLKVLGMVEGKGAPEGALYL
Rv3568c|M.tuberculosis_H37Rv -----MS---IRSLGYLRIEATDMAAWREYGLKVLGMVEGKGAPEGALYL
MSMEG_2891|M.smegmatis_MC2_155 ---------MIKSLGYVTVQATDMQRWRHFAFGVLGFAEGKGPDPDALYL
TH_4499|M.thermoresistible__bu MRHERDTMGFIKSLGYVIVQATDMDRWRHFAYEVLGFAEGTGPNPEALYL
MAB_0076|M.abscessus_ATCC_1997 ---------MLTSLGYLTVATRDMDRWRHFALRVLGFAEGSGPDPDALYL
Mflv_0538|M.gilvum_PYR-GCK --------MLVKTLGYVGVESPDAKEWLAFGPEVLGMEAVEASS-GSVLL
Mvan_4413|M.vanbaalenii_PYR-1 -------MNRVVGLGYLIIEATDLEEWQTFACDLLGLQAVVRTP-DRLSL
: ***: : : * * :. :**: : *
MMAR_5063|M.marinum_M RMDDFPARLVIVPGTQDRLAEAGWECADAQGLQEIRNRLDVEGTPYKEAT
MUL_4139|M.ulcerans_Agy99 RMDDFPARLVIVPGTQDRLAEAGWECADAQGLQEIRNRLDVEGTPYKEAT
MAV_0593|M.avium_104 RMDDFPARLVIVPGERDHLLEAGWECANAEGLQEIRDRLDMEGVPYKEAT
Mb3599c|M.bovis_AF2122/97 RMDDFPARLVVVPGEHDRLLEAGWECANAEGLQEIRNRLDLEGTPYKEAT
Rv3568c|M.tuberculosis_H37Rv RMDDFPARLVVVPGEHDRLLEAGWECANAEGLQEIRNRLDLEGTPYKEAT
MSMEG_2891|M.smegmatis_MC2_155 RMDERCARIIVTPGDVDKIVTVGWEVRDHAALQEVKRALDAAGVPHKQLS
TH_4499|M.thermoresistible__bu RMDERTARITVEPGEVDRIVTIGWEVRDHAALEDVKRALAAAGVPHRQLS
MAB_0076|M.abscessus_ATCC_1997 RMDERAARVVVVPGETDQVITVGWEVRDGAALRAVVRALEAADIGVERLS
Mflv_0538|M.gilvum_PYR-GCK RIDDADHRLAVHHGERNRMLYAGWDVGSEEALEAAGELLHKRGIGFEVGT
Mvan_4413|M.vanbaalenii_PYR-1 RVDEKAYRIDVRQAESDAVAVIGWEVRGPRELGQLANLLQANGYDVKWAG
*:*: *: : . : : **: . * * . .
MMAR_5063|M.marinum_M AAELADRRVDEMILFSDPSGNPLAVFHGAALEHRRVVSPYGHK-FVTGEQ
MUL_4139|M.ulcerans_Agy99 AAELADRRVDEMILFSDPSGNPLAVFHGAALEHRRVVSPYGHK-FVTGEQ
MAV_0593|M.avium_104 AAQLADRRVVEMIRFSDPSGNCLEVFHGVALEHRRVVSPYGHK-FVTGEQ
Mb3599c|M.bovis_AF2122/97 AAELADRRVDEMIRFADPSGNCLEVFHGTALEHRRVVSPYGHR-FVTGEQ
Rv3568c|M.tuberculosis_H37Rv AAELADRRVDEMIRFADPSGNCLEVFHGTALEHRRVVSPYGHR-FVTGEQ
MSMEG_2891|M.smegmatis_MC2_155 LAEADSRRVEEAIAFEDPAGTSLEVFHGPVLDHSPVVTPFGAR-FVTGDQ
TH_4499|M.thermoresistible__bu TAEADARRVEEVVAFADPAGTALEVFHGPVLDHSPVVTPFGAR-FVTGDQ
MAB_0076|M.abscessus_ATCC_1997 PEEADSRRAEEVVAFTDPAGTPVEVFHGPVLDHSPVVTPYGAR-FVTGAQ
Mflv_0538|M.gilvum_PYR-GCK EEDCAARGVLGFLTLSDPSGLRHELFYGQKVVPGSFRPGRAISGFVTGAQ
Mvan_4413|M.vanbaalenii_PYR-1 AEVARERRVSGLAGFTDPDGQAVELFWGLQEHRDRFVSPTGAR-FVTGAA
* . : ** * :* * . . . ****
MMAR_5063|M.marinum_M GLGHVVLTTRDDAEALHFYRDVLGFKLRDSMRMPPQVVGRPADGAPAWLR
MUL_4139|M.ulcerans_Agy99 GLGHVVLTTRDDAEALHFYRDVLGFKLRDSMRMPPQVVGRPADGAPAWLR
MAV_0593|M.avium_104 GLGHVVLSTRDDEESLHFYRDVLGFKLRDSMRMPPQVVGRPADGAPAWLR
Mb3599c|M.bovis_AF2122/97 GMGHVVLSTRDDAEALHFYRDVLGFRLRDSMRLPPRMVGRPADGPPAWLR
Rv3568c|M.tuberculosis_H37Rv GMGHVVLSTRDDAEALHFYRDVLGFRLRDSMRLPPQMVGRPADGPPAWLR
MSMEG_2891|M.smegmatis_MC2_155 GLGHVVLPALDVNGLFEFYTETLGFKSRGAFRVP-----VPPEFGPVRVR
TH_4499|M.thermoresistible__bu GLGHVVLPTVDVDGLFEFYTEVLGFRSRGAFRVP-----APEEFGPVRVR
MAB_0076|M.abscessus_ATCC_1997 GLGHVVLPVTDPEGAFTFYTEVLGFRPRGAFRIP-----APPEFGPMRVR
Mflv_0538|M.gilvum_PYR-GCK GLGHVVLATPDLAQADRFLRGVLGFKKSD------------EIYTFMDLW
Mvan_4413|M.vanbaalenii_PYR-1 GLGHVFQAVDDADVYSELYQNLLGFKLSDIIDFG----------PGASAT
*:***. .. * : ***: .
MMAR_5063|M.marinum_M FFGCNPRHHSLAFLPLPTP--SGIVHLMVEVQNSDDVGLCLDRALRRKVP
MUL_4139|M.ulcerans_Agy99 FFGCNPRHHSLAFLPLPTP--SGIVHLMVEVQNSDDVGLCLDRALRRKVP
MAV_0593|M.avium_104 FFGCNPRHHSLAFLPLPTP--SGIVHLMLEVENADDVGLCLDRALRRKVP
Mb3599c|M.bovis_AF2122/97 FFGCNPRHHSLAFLPMPTS--SGIVHLMVEVEQADDVGLCLDRALRRKVP
Rv3568c|M.tuberculosis_H37Rv FFGCNPRHHSLAFLPMPTS--SGIVHLMVEVEQADDVGLCLDRALRRKVP
MSMEG_2891|M.smegmatis_MC2_155 FLGVNERHHSLALCPASTLRDPGLVHLMVEVDSLDAVGQALDRVNADGFQ
TH_4499|M.thermoresistible__bu FLGINERHHSLALCPASTLRDPGLIHLMVEVDTLDAVGQALDRVNRDGYQ
MAB_0076|M.abscessus_ATCC_1997 FLGINERHHSLALCPAPHGGAPGLVHVMVEVDSLDAVGQALDRVLKDDYS
Mflv_0538|M.gilvum_PYR-GCK FYHCNPRHHSIALTPMPGVR--GLHHVMVEVQSFDDVGMAYDLCMSRNIP
Mvan_4413|M.vanbaalenii_PYR-1 FLHTTERHHSYAFAALPGLP-NGVGHVMLEVDDLDIVGRAWEKVRDGAAP
* . **** *: . . *: *:*:**: * ** . :
MMAR_5063|M.marinum_M MSATLGRHVNDLMLSFYMKTPGGFDVEFGCEGRQVDDHDWIARESTAVSL
MUL_4139|M.ulcerans_Agy99 MSATLGRHVNDLMLSFYMKTPGGFDVEFGCEGRQVDDHDWIARESTAVSL
MAV_0593|M.avium_104 MSATLGRHVNDLMLSFYMKTPGGFDVEFGCEGRQVEDDDWVARESTAVSL
Mb3599c|M.bovis_AF2122/97 MSATLGRHVNDLMLSFYMKTPGGFDIEFGCEGRQVDDRDWIARESTAVSL
Rv3568c|M.tuberculosis_H37Rv MSATLGRHVNDLMLSFYMKTPGGFDIEFGCEGRQVDDRDWIARESTAVSL
MSMEG_2891|M.smegmatis_MC2_155 LSSTLGRHTNDKMVSFYVRAPGDWDIEFGTDGMRVDEQHYTAEEITADSY
TH_4499|M.thermoresistible__bu LSSTLGRHTNDKMVSFYVRAPGDWDIEFGTAGMRVDETYYTAEEITADSY
MAB_0076|M.abscessus_ATCC_1997 LSSTLGRHTNDKMVSFYVRAPGGWDIEFGTDGMRVDETCYTAEEITADSY
Mflv_0538|M.gilvum_PYR-GCK LSMTLGRHVNDRMVSFYVRTPSGFDIEYGWDAVTVDEETWTVAQYDRPSV
Mvan_4413|M.vanbaalenii_PYR-1 MVLDFGKHTNDEMLSFYVKTPSGFELEYGYGGKTIDDATWTPSRYDAASY
: :*:*.** *:***:::*..:::*:* . ::: : . *
MMAR_5063|M.marinum_M WGHDFTVGARG--------
MUL_4139|M.ulcerans_Agy99 WGHDFTVGARG--------
MAV_0593|M.avium_104 WGHDFTVGMRG--------
Mb3599c|M.bovis_AF2122/97 WGHDFTVGARG--------
Rv3568c|M.tuberculosis_H37Rv WGHDFTVGARG--------
MSMEG_2891|M.smegmatis_MC2_155 WGHQWVSDLPRAMQP----
TH_4499|M.thermoresistible__bu WGHQWVGDLPAAMRP----
MAB_0076|M.abscessus_ATCC_1997 WGHDWSGSQPLAAM-----
Mflv_0538|M.gilvum_PYR-GCK WGHQMVAQTPPGALEAATT
Mvan_4413|M.vanbaalenii_PYR-1 WGHRRTRPTDPDSAELSV-
***