For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLVKALGYVGVESPDAKEWLAFGPEVLGMEAVEASSGSVLLRIDDADHRLAVHHGERNRMLYAGWDVGSE EALEAAGELLHKRGIGFEVGTEEDCAARGVLGFVTLSDPSGLRHELFYGQKVVPGSFRPGRAISGFVTGA QGLGHVVLATPDLAQADRFLRGVLGFKKSDEIYTFMDLWFYHCNPRHHSIALTPMPGVRGLHHVMVEVQS FDDVGMAYDLCMSRNIPLSMTLGRHVNDRMVSFYVRTPSGFDIEYGWDAVTVDEETWTVAQYDRPSVWGH QMVAQTPPGALEAATT
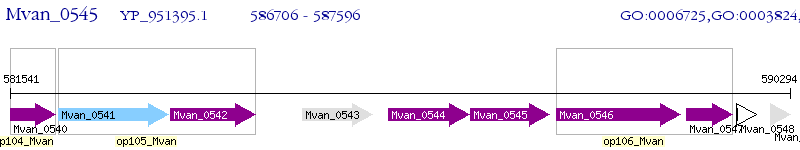
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0545 | - | - | 100% (296) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. vanbaalenii PYR-1 | Mvan_0470 | - | e-175 | 98.99% (296) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. vanbaalenii PYR-1 | Mvan_5306 | - | 2e-53 | 39.32% (295) | glyoxalase/bleomycin resistance protein/dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3599c | bphC | 8e-58 | 41.08% (297) | putative biphenyl-2,3-diol 1,2-dioxygenase BPHC (23OHBP |
| M. gilvum PYR-GCK | Mflv_0538 | - | 1e-175 | 99.32% (296) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | Rv3568c | bphC | 5e-58 | 41.08% (297) | biphenyl-2,3-diol 1,2-dioxygenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0585 | - | 2e-59 | 40.75% (292) | 2,3-dihydroxybiphenyl 1,2-dioxygenase |
| M. marinum M | MMAR_5063 | bphC | 3e-56 | 39.73% (297) | biphenyl-2,3-diol 1,2-dioxygenase BphC |
| M. avium 104 | MAV_0593 | - | 5e-57 | 39.32% (295) | extradiol ring-cleavage dioxygenase |
| M. smegmatis MC2 155 | MSMEG_6036 | - | 4e-52 | 38.05% (297) | biphenyl-2,3-diol 1,2-dioxygenase |
| M. thermoresistible (build 8) | TH_1740 | - | 5e-58 | 40.07% (297) | PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC (23OHBP |
| M. ulcerans Agy99 | MUL_4139 | bphC | 2e-56 | 39.73% (297) | biphenyl-2,3-diol 1,2-dioxygenase BphC |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5063|M.marinum_M MS---IRSLGYLRIEATDMAAWREYGLKVLGMVEGKGSTEGALYLRMDDF
MUL_4139|M.ulcerans_Agy99 MS---IRSLGYLRIEATDMAAWREYGLKVLGMVEGKGSTEGALYLRMDDF
MAV_0593|M.avium_104 MSNGAIRSLGYLRIEATDMAAWRDYGLKVLGMVEGKGDTEGALYLRMDDF
Mb3599c|M.bovis_AF2122/97 MS---IRSLGYLRIEATDMAAWREYGLKVLGMVEGKGAPEGALYLRMDDF
Rv3568c|M.tuberculosis_H37Rv MS---IRSLGYLRIEATDMAAWREYGLKVLGMVEGKGAPEGALYLRMDDF
MSMEG_6036|M.smegmatis_MC2_155 MS---IKSLGYLRIEATDVAAWREYGLKVLGMVEGKGNTDGALYLRMDEF
TH_1740|M.thermoresistible__bu MS---IRSLGYLRIEATDVAAWREYGLKVLGMVEGKGPTEGALYLRMDEF
MAB_0585|M.abscessus_ATCC_1997 MS--IIKALGYMRIQATDVAAWREFGTKVLGMMEGTGDDPAALYLRMDDF
Mvan_0545|M.vanbaalenii_PYR-1 ML---VKALGYVGVESPDAKEWLAFGPEVLGMEAVEASS-GSVLLRIDDA
Mflv_0538|M.gilvum_PYR-GCK ML---VKTLGYVGVESPDAKEWLAFGPEVLGMEAVEASS-GSVLLRIDDA
* :::***: :::.* * :* :**** . .:: **:*:
MMAR_5063|M.marinum_M PARLVIVPGTQDRLAEAGWECADAQGLQEIRNRLDVEGTPYKEATAAELA
MUL_4139|M.ulcerans_Agy99 PARLVIVPGTQDRLAEAGWECADAQGLQEIRNRLDVEGTPYKEATAAELA
MAV_0593|M.avium_104 PARLVIVPGERDHLLEAGWECANAEGLQEIRDRLDMEGVPYKEATAAQLA
Mb3599c|M.bovis_AF2122/97 PARLVVVPGEHDRLLEAGWECANAEGLQEIRNRLDLEGTPYKEATAAELA
Rv3568c|M.tuberculosis_H37Rv PARLVVVPGEHDRLLEAGWECANAEGLQEIRNRLDLEGTPYKEATAAELA
MSMEG_6036|M.smegmatis_MC2_155 PARLVIVPGEHDRLQVSGWEAANAAELQDIRSRLDAAGTPFKEGTSAELA
TH_1740|M.thermoresistible__bu PARLVIVPGERDRLLESGWECANAEGLQDVRSRLEAAGVPYKEGTAAELA
MAB_0585|M.abscessus_ATCC_1997 AARLVIVPGESDRLLASGWEVADAPALQAVRERLAKAGVEYAEGDKAEVS
Mvan_0545|M.vanbaalenii_PYR-1 DHRLAVHHGERNRMLYAGWDVGSEEALEAAGELLHKRGIGFEVGTEEDCA
Mflv_0538|M.gilvum_PYR-GCK DHRLAVHHGERNRMLYAGWDVGSEEALEAAGELLHKRGIGFEVGTEEDCA
**.: * ::: :**: .. *: . * * : . : :
MMAR_5063|M.marinum_M DRRVDEMILFSDPSGNPLAVFHGAALEHRRVVSPYGHK-FVTGEQGLGHV
MUL_4139|M.ulcerans_Agy99 DRRVDEMILFSDPSGNPLAVFHGAALEHRRVVSPYGHK-FVTGEQGLGHV
MAV_0593|M.avium_104 DRRVVEMIRFSDPSGNCLEVFHGVALEHRRVVSPYGHK-FVTGEQGLGHV
Mb3599c|M.bovis_AF2122/97 DRRVDEMIRFADPSGNCLEVFHGTALEHRRVVSPYGHR-FVTGEQGMGHV
Rv3568c|M.tuberculosis_H37Rv DRRVDEMIRFADPSGNCLEVFHGTALEHRRVVSPYGHR-FVTGEQGMGHV
MSMEG_6036|M.smegmatis_MC2_155 ERRVDEMIVFDDPSGNTLEVFHGVALEHRRVVSPYGHK-FVTEEQGLGHV
TH_1740|M.thermoresistible__bu DRRVVELIRFQDPSGNTLEAFHTVALEHRRVVSPYGHR-FVTGEQGLGHV
MAB_0585|M.abscessus_ATCC_1997 DRKVEAFIQFSDPSGNTLEVFHGAQYMGRRFVSPYGHK-FVTAEQGLGHV
Mvan_0545|M.vanbaalenii_PYR-1 ARGVLGFVTLSDPSGLRHELFYGQKVVPGSFRPGRAISGFVTGAQGLGHV
Mflv_0538|M.gilvum_PYR-GCK ARGVLGFLTLSDPSGLRHELFYGQKVVPGSFRPGRAISGFVTGAQGLGHV
* * :: : **** *: . . . *** **:***
MMAR_5063|M.marinum_M VLTTRDDAEALHFYRDVLGFKLRDSMRMPPQVVGRPADGAPAWLRFFGCN
MUL_4139|M.ulcerans_Agy99 VLTTRDDAEALHFYRDVLGFKLRDSMRMPPQVVGRPADGAPAWLRFFGCN
MAV_0593|M.avium_104 VLSTRDDEESLHFYRDVLGFKLRDSMRMPPQVVGRPADGAPAWLRFFGCN
Mb3599c|M.bovis_AF2122/97 VLSTRDDAEALHFYRDVLGFRLRDSMRLPPRMVGRPADGPPAWLRFFGCN
Rv3568c|M.tuberculosis_H37Rv VLSTRDDAEALHFYRDVLGFRLRDSMRLPPQMVGRPADGPPAWLRFFGCN
MSMEG_6036|M.smegmatis_MC2_155 VLTTKDDRETLHFYRDILGFSLRDSMKLPPQLVGRPADGEPAWLRFLGVN
TH_1740|M.thermoresistible__bu VLTTRDDAESLRFYRDVLGFYLRDSMRLPPQLVGRPADGEPAWLRFFGCN
MAB_0585|M.abscessus_ATCC_1997 VLTCTDDVAAQEFYQDVLGFQLRDSMKLPPQLVGRPADGDPVWLRFYGCN
Mvan_0545|M.vanbaalenii_PYR-1 VLATPDLAQADRFLRGVLGFKKSDEIYTFMD------------LWFYHCN
Mflv_0538|M.gilvum_PYR-GCK VLATPDLAQADRFLRGVLGFKKSDEIYTFMD------------LWFYHCN
**: * : .* :.:*** *.: * * *
MMAR_5063|M.marinum_M PRHHSLAFLPLPTPSGIVHLMVEVQNSDDVGLCLDRALRRKVPMSATLGR
MUL_4139|M.ulcerans_Agy99 PRHHSLAFLPLPTPSGIVHLMVEVQNSDDVGLCLDRALRRKVPMSATLGR
MAV_0593|M.avium_104 PRHHSLAFLPLPTPSGIVHLMLEVENADDVGLCLDRALRRKVPMSATLGR
Mb3599c|M.bovis_AF2122/97 PRHHSLAFLPMPTSSGIVHLMVEVEQADDVGLCLDRALRRKVPMSATLGR
Rv3568c|M.tuberculosis_H37Rv PRHHSLAFLPMPTSSGIVHLMVEVEQADDVGLCLDRALRRKVPMSATLGR
MSMEG_6036|M.smegmatis_MC2_155 PRHHSLAFMPGQTPSGIVHLMVEVENSDDVGLCLDRALRRKVKMSATLGR
TH_1740|M.thermoresistible__bu PRHHSLAFMPGETPSGIVHLMVEVENSDDVGLCLDRALRRKVPMSATLGR
MAB_0585|M.abscessus_ATCC_1997 PRHHALAFMPMPNPTGIVHLMVEVENSDDVGLCLDRANRRKVKMSATLGR
Mvan_0545|M.vanbaalenii_PYR-1 PRHHSIALTPMPGVRGLHHVMVEVQSFDDVGMAYDLCMSRNIPLSMTLGR
Mflv_0538|M.gilvum_PYR-GCK PRHHSIALTPMPGVRGLHHVMVEVQSFDDVGMAYDLCMSRNIPLSMTLGR
****::*: * *: *:*:**:. ****:. * . *:: :* ****
MMAR_5063|M.marinum_M HVNDLMLSFYMKTPGGFDVEFGCEGRQVDDHDWIARESTAVSLWGHDFTV
MUL_4139|M.ulcerans_Agy99 HVNDLMLSFYMKTPGGFDVEFGCEGRQVDDHDWIARESTAVSLWGHDFTV
MAV_0593|M.avium_104 HVNDLMLSFYMKTPGGFDVEFGCEGRQVEDDDWVARESTAVSLWGHDFTV
Mb3599c|M.bovis_AF2122/97 HVNDLMLSFYMKTPGGFDIEFGCEGRQVDDRDWIARESTAVSLWGHDFTV
Rv3568c|M.tuberculosis_H37Rv HVNDLMLSFYMKTPGGFDIEFGCEGRQVDDRDWIARESTAVSLWGHDFTV
MSMEG_6036|M.smegmatis_MC2_155 HVNDKMLSFYMKTPGGFDIEFGCEGLEVDDDNWVARESTAVSLWGHDFSV
TH_1740|M.thermoresistible__bu HVNDKMLSFYMKTPGGFDVEFGCEGLQVDDEEWVARESTAVSLWGHDFSI
MAB_0585|M.abscessus_ATCC_1997 HINDKMLSFYMKTPGGFDMEFGCEGMEVDDESWVARESVGVSLWGHDFSV
Mvan_0545|M.vanbaalenii_PYR-1 HVNDRMVSFYVRTPSGFDIEYGWDAVTVDEETWTVAQYDRPSVWGHQMVA
Mflv_0538|M.gilvum_PYR-GCK HVNDRMVSFYVRTPSGFDIEYGWDAVTVDEETWTVAQYDRPSVWGHQMVA
*:** *:***::**.***:*:* :. *:: * . : *:***::
MMAR_5063|M.marinum_M GARG--------
MUL_4139|M.ulcerans_Agy99 GARG--------
MAV_0593|M.avium_104 GMRG--------
Mb3599c|M.bovis_AF2122/97 GARG--------
Rv3568c|M.tuberculosis_H37Rv GARG--------
MSMEG_6036|M.smegmatis_MC2_155 GFK---------
TH_1740|M.thermoresistible__bu GFKG--------
MAB_0585|M.abscessus_ATCC_1997 GFK---------
Mvan_0545|M.vanbaalenii_PYR-1 QTPPGALEAATT
Mflv_0538|M.gilvum_PYR-GCK QTPPGALEAATT