For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLHGKVAFVAGASRGIGATVAAALARAGASVAVAARSEQEGKLPGTIGSVAQGISAAGGRALPVACDVT SEESVNAAVAETVAEFGGIDILVANAGVLWLGPIETTPLKRWQLCLDVNTTGVFLVTKAVIPHVRARGGG SLIAVTTTGVNMTDHGANAYWVSKAAVERLYLGLAADLKPDNIAVNCLSPSRVVLTEGWQAGGAGMQIPP EMVEPPEAMADAAVFLAGQDASGITGTIQRSEALTF
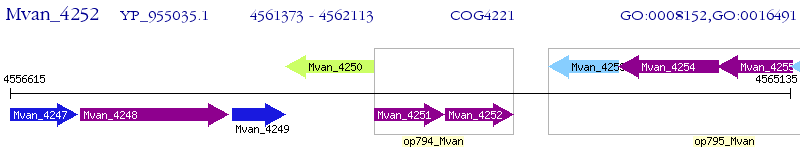
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4252 | - | - | 100% (246) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4262 | - | 2e-33 | 34.32% (236) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1776 | - | 1e-32 | 37.33% (225) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3251 | - | 1e-35 | 36.55% (238) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2396 | - | 1e-123 | 90.98% (244) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3224 | - | 1e-35 | 36.55% (238) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-15 | 29.96% (247) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_3946c | - | 2e-31 | 39.20% (199) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_1333 | - | 1e-35 | 36.13% (238) | iron-regulated short-chain dehydrogenase/reductase |
| M. avium 104 | MAV_4172 | - | 1e-36 | 37.08% (240) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4872 | - | 1e-112 | 81.56% (244) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_0664 | - | 2e-34 | 36.59% (246) | POSSIBLE IRON-REGULATED SHORT-CHAIN DEHYDROGENASE/REDUCTASE |
| M. ulcerans Agy99 | MUL_2546 | - | 7e-36 | 36.13% (238) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4252|M.vanbaalenii_PYR-1 ----------MTLHGKVAFVAGASRGIGATVAAALARAGASVAVAARSEQ
Mflv_2396|M.gilvum_PYR-GCK ----------MTLDGKVAFVAGASRGIGATVAAALARAGASVAVAARTEQ
MSMEG_4872|M.smegmatis_MC2_155 MRETPKPSTPGSLAGKVAFVAGASRGIGATIATALAAEGAAVAVAARSER
Mb3251|M.bovis_AF2122/97 ----------MSLNGKTMFISGASRGIGLAIAKRAARDGANIALIAKTAE
Rv3224|M.tuberculosis_H37Rv ----------MSLNGKTMFISGASRGIGLAIAKRAARDGANIALIAKTAE
MMAR_1333|M.marinum_M ----------MSLNGKTMFISGASRGIGLAIAKRAAQDGANIALIAKTAE
MUL_2546|M.ulcerans_Agy99 ----------MSLNGKTMFISGASRGIGLAIAKRAAQDGANIALIAKTAE
MAV_4172|M.avium_104 ----------MSLNGKTMFISGASRGIGLAIAKRAAQDGANIALIAKTAE
TH_0664|M.thermoresistible__bu ----------MSLRGKTMFISGGSRGIGLAIAKRVAADGANVALVAKSAE
MAB_3946c|M.abscessus_ATCC_199 ---------MGSLAGKTIIMSGGSRGIGLAIALRAARDGANVAIMAKTAE
MLBr_01807|M.leprae_Br4923 -----------MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVAR
. . . :. .** :: *. .: .
Mvan_4252|M.vanbaalenii_PYR-1 EG-KLPGTIGSVAQGISAAGGRALPVACDVTSEESVNAAVAETVAEFGGI
Mflv_2396|M.gilvum_PYR-GCK EG-RLPGTIGSVAQRITDEGGRALPVVCDVTREESVEAAVAATVAEFGGI
MSMEG_4872|M.smegmatis_MC2_155 PG-KLPGTIDSVASAITDAGGHALPVVCDVTDEQSVERAVAQTVSELGGI
Mb3251|M.bovis_AF2122/97 PHPKLPGTVFTAAKELEEAGGQALPIVGDIRDPDAVASAVATTVEQFGGI
Rv3224|M.tuberculosis_H37Rv PHPKLPGTVFTAAKELEEAGGQALPIVGDIRDPDAVASAVATTVEQFGGI
MMAR_1333|M.marinum_M PHPKLPGTVYTAAKELEEAGGQALPIVGDVRDPDSVSAAVAKTVEQFGGI
MUL_2546|M.ulcerans_Agy99 PHPKLPGTVYTAAKELEEAGGQALPIVGDVRDPDSVSAAVAKTVEQFGGI
MAV_4172|M.avium_104 PHPKLPGTVYTAAKELEEAGGQALPIVGDVRDPESVEAAVAKTVEQFGGI
TH_0664|M.thermoresistible__bu PHPKLPGTIYTAAKEIEEAGGQALPIVGDIRDGDAVAAAVAKTVEQFGGI
MAB_3946c|M.abscessus_ATCC_199 PHPKLPGTIYTAAEEIEAAGGHALPLLGDVRDDEVVASAVAQTVEKFGGI
MLBr_01807|M.leprae_Br4923 RL-VADGHRVAVTHRASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQGPV
* :.: . : : *: : : *. . . * :
Mvan_4252|M.vanbaalenii_PYR-1 DILVANAGVLWLGPIETTPLKRWQLCLDVNTTGVFLVTKAVIP-HVRARG
Mflv_2396|M.gilvum_PYR-GCK DILVANAGVLWLGPIESTPLKRWQLCLDVNTTGVFLVTKAVIP-HVRARG
MSMEG_4872|M.smegmatis_MC2_155 DILVANAGVLWLGPIENTPLKRWQLCLDVNLTGVFLVTKAVIP-HVRARG
Mb3251|M.bovis_AF2122/97 DICVNNASAINLGSITEVPMKRFDLMNGIQVRGTYAVSQACIP-HMKGRE
Rv3224|M.tuberculosis_H37Rv DICVNNASAINLGSITEVPMKRFDLMNGIQVRGTYAVSQACIP-HMKGRE
MMAR_1333|M.marinum_M DICVNNASAINLGSITEVPMKRFDLMNGIQVRGTYAVSQACIP-HLKGRE
MUL_2546|M.ulcerans_Agy99 DICVNNASAINLGSITEVPMKRFDLMNGIQVRGTYAVSQACIP-HLKGRE
MAV_4172|M.avium_104 DICVNNASAINLGPITEVPMKRFDLMNGIQVRGTYAVSQACIP-HLKGRE
TH_0664|M.thermoresistible__bu DICVNNASAINLGSIEEVPLKRFDLMNGIQVRGTYAVSQSCIP-HMKGRD
MAB_3946c|M.abscessus_ATCC_199 DIVVNNASALNLAPSESISMKAYDLMQDINARGSFSLSTSAIG-ALKEAD
MLBr_01807|M.leprae_Br4923 EVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQRF
:: * **. . : :: .:: * : :: ::
Mvan_4252|M.vanbaalenii_PYR-1 GGSLIAVTTTGVNMTDHGAN----AYWVSKAAVERLYLGLAADLKPDNIA
Mflv_2396|M.gilvum_PYR-GCK GGSLIAVTTTGVTMTDHGAN----AYWVSKAAAERLYLGLAADLKPDNIA
MSMEG_4872|M.smegmatis_MC2_155 GGSLIAITTTGVDMVEHGSN----AYWVSKAATERLYTGLAADLRDDNIA
Mb3251|M.bovis_AF2122/97 NPHILTLSPP-ILLEKKWLR--PTAYMMAKYGMTLCALGIAEEMRADGIA
Rv3224|M.tuberculosis_H37Rv NPHILTLSPP-ILLEKKWLR--PTAYMMAKYGMTLCALGIAEEMRADGIA
MMAR_1333|M.marinum_M NPHILTLSPP-VQLDKKWLK--PTAYMMAKFGMTLCALGIAEEMRDEGIA
MUL_2546|M.ulcerans_Agy99 NPHILTLSPP-VQLDKKWLK--PTAYMMAKFGMTLCALGIAEEMRDEGIA
MAV_4172|M.avium_104 NPHILTLSPP-VLLGKEWLK--PTAYMMAKFGMTLCALGIAEEMREAGIA
TH_0664|M.thermoresistible__bu NPHILTLSPP-IRLEPKWLR--PTPYMMAKYGMTLCALGIAEELRDAGIA
MAB_3946c|M.abscessus_ATCC_199 NPHILTLSPP-ITLEPKWFERTTTAYTISKFSMSLVAIGLAAELRQYGIA
MLBr_01807|M.leprae_Br4923 GRYIFMGSVVGLSGVPGQAN-----YAASKSGLIGMARSLARELGSRSIT
. :: : : . * :* . .:* :: .*:
Mvan_4252|M.vanbaalenii_PYR-1 VNCLSPSRVVLTEGWQAGGAGMQIPPEMVEPP------EAMADAAVFLAG
Mflv_2396|M.gilvum_PYR-GCK VNCLSPSRVVLTEGWQAGGAGMEIPPEMVEPP------EAMGRATVMLAQ
MSMEG_4872|M.smegmatis_MC2_155 VNCLSPSRVVLTEGWQAGGGGLKIPPEMIEPP------EAMARAAVLLAQ
Mb3251|M.bovis_AF2122/97 SNTLWPRTMVATAAVQNLLGGDEAMARSRKP-------EVYADAAYVIVN
Rv3224|M.tuberculosis_H37Rv SNTLWPRTMVATAAVQNLLGGDEAMARSRKP-------EVYADAAYVIVN
MMAR_1333|M.marinum_M SNTLWPRTLVATAAVQNLLGGDEAMGRARKP-------DVYADAAYVILN
MUL_2546|M.ulcerans_Agy99 SNTLWPRTLVATAAVQNLLGGEEAMGRARKP-------DVYADAAYVILN
MAV_4172|M.avium_104 SNTLWPRTLVATAAVQNLLGGDEAMGRARKP-------EVYSDAAYVILN
TH_0664|M.thermoresistible__bu SNTLWPRTTVATAAVQNLLGGDEAMARSRKP-------EVYADAAYVVLN
MAB_3946c|M.abscessus_ATCC_199 SNALWPRTTIDTAAIRNILG-AELVARCRSA-------QIMADAAYEILT
MLBr_01807|M.leprae_Br4923 ANVVAPG-FIDTEMTRAMDSKYQEAALQAIPLGRIGAVEEIAGAVSFLAS
* : * : * : . : . : . *. :
Mvan_4252|M.vanbaalenii_PYR-1 QDASGITGTIQRSEALTF--------------------------------
Mflv_2396|M.gilvum_PYR-GCK QDASGVTGTIQRSEALAL--------------------------------
MSMEG_4872|M.smegmatis_MC2_155 QDGTGVTGTVQRSEALTL--------------------------------
Mb3251|M.bovis_AF2122/97 KPATEYTGKTLLCEDVLVESGVTDLSVYDCVPGATLGVDLWVEDANPPGY
Rv3224|M.tuberculosis_H37Rv KPATEYTGKTLLCEDVLVESGVTDLSVYDCVPGATLGVDLWVEDANPPGY
MMAR_1333|M.marinum_M KNSREYTGNSALCEDVLLESGVSDLSIYNCVPGAKLGVDLWVEEVNPPGY
MUL_2546|M.ulcerans_Agy99 KNSREYTGNSALCEDVLLESGVSDLSIYNCVPGAKLGVDLWVEEVNPPGY
MAV_4172|M.avium_104 KPAREFTGNMLLCEDVLVESGVTDLSVYNCVPGSQLGVDFWVDGVNPPGY
TH_0664|M.thermoresistible__bu KPSS-YTGNTLLCEDVLLESGVTDLSVYDCVPGSELGVDLWVDSPNPPGY
MAB_3946c|M.abscessus_ATCC_199 KPSREVSGNCFIDDEVLREAGVTDFSQYRECAEEDLELDFWMERA-----
MLBr_01807|M.leprae_Br4923 EDAGYISGAVIPVDGGLGMGH-----------------------------
: . :* :
Mvan_4252|M.vanbaalenii_PYR-1 ---
Mflv_2396|M.gilvum_PYR-GCK ---
MSMEG_4872|M.smegmatis_MC2_155 ---
Mb3251|M.bovis_AF2122/97 LPA
Rv3224|M.tuberculosis_H37Rv LPA
MMAR_1333|M.marinum_M TQP
MUL_2546|M.ulcerans_Agy99 TQP
MAV_4172|M.avium_104 TGP
TH_0664|M.thermoresistible__bu TGP
MAB_3946c|M.abscessus_ATCC_199 ---
MLBr_01807|M.leprae_Br4923 ---