For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQIAGSSALVVGGAGGLGEATVRRLHDAGAKVVIADLADDKGRQLESELGVRYVRTDATSEESVNAAITE AESLGPLRISVDTHGGPAGGGRLVGKDGSPLSMEAFETTIKFYLTAVFNIMRLSAAAIAKTEPLEEGGRG VIVNTASVAGYEGQIGQLPYSAAKGGVIGMTLVAARDLSPLGIRVVTIAPGTINTPAYGKAADQLEQYWG PQVPFPKRMGRSTEYAQLAQSVIENDYLNGEVIRLDGALRFPPK
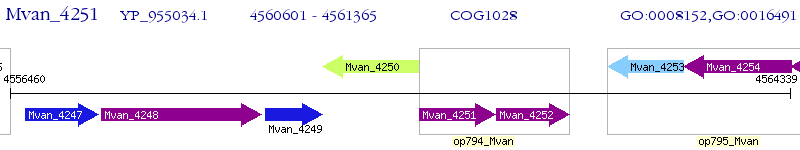
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4251 | - | - | 100% (254) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4577 | - | 7e-59 | 49.22% (256) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_2027 | - | 2e-50 | 42.41% (257) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1176 | - | 2e-56 | 48.05% (256) | short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_2127 | - | 2e-58 | 49.22% (256) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1144 | - | 2e-56 | 48.05% (256) | short-chain type dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-15 | 30.49% (246) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0368 | - | 3e-64 | 51.15% (260) | Short-chain dehydrogenase/reductas |
| M. marinum M | MMAR_4307 | - | 4e-56 | 47.29% (258) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2552 | - | 1e-74 | 57.94% (252) | 3-hydroxyacyl-CoA dehydrogenase type-2 |
| M. smegmatis MC2 155 | MSMEG_4871 | - | 1e-128 | 88.19% (254) | 3-hydroxyacyl-CoA dehydrogenase type-2 |
| M. thermoresistible (build 8) | TH_3346 | - | 1e-129 | 89.76% (254) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0987 | - | 6e-57 | 47.67% (258) | short-chain type dehydrogenase/reductase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4871|M.smegmatis_MC2_155 -----MQIA-----------GTSALVVGGTGGLGEATVRRLHGAGAKVVV
TH_3346|M.thermoresistible__bu -----MQIA-----------GSAALVVGGAGGLGEATVRRLHAAGAKVVV
Mvan_4251|M.vanbaalenii_PYR-1 -----MQIA-----------GSSALVVGGAGGLGEATVRRLHDAGAKVVI
MAV_2552|M.avium_104 -----MTVAKFD--------GASAIVSGGAGGLGEATVRRLHADGLGVVI
MMAR_4307|M.marinum_M -----MEIR-----------DAVAVVTGGASGLGLATTKRLLDAGAQVVV
MUL_0987|M.ulcerans_Agy99 -----MEIR-----------DAVAVVTGGASGLGLATTKRLLDAGAQVVV
Mb1176|M.bovis_AF2122/97 -----MKTK-----------DAVAVVTGGASGLGLATTKRLLDAGAQVVV
Rv1144|M.tuberculosis_H37Rv -----MKTK-----------DAVAVVTGGASGLGLATTKRLLDAGAQVVV
Mflv_2127|M.gilvum_PYR-GCK -----MEIK-----------DAAAVVTGGASGLGLATTKRLLDRGASVVV
MAB_0368|M.abscessus_ATCC_1997 -----MDIN-----------GVSAIVTGGASGLGAATARLLAAQGAKVVI
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
. :* ** *:* *..: * * *.:
MSMEG_4871|M.smegmatis_MC2_155 ADVADDKGAALEKELG--IRYVRTDATSEESVLAAIAEAESLA-PLRISV
TH_3346|M.thermoresistible__bu ADLADERGGELEKELG--VRYVRTDATSEQDVLAAIAEAESLG-PLRISV
Mvan_4251|M.vanbaalenii_PYR-1 ADLADDKGRQLESELG--VRYVRTDATSEESVNAAITEAESLG-PLRISV
MAV_2552|M.avium_104 ADLAADKGKALADELGSGALFVSTDVTSDESVQGAIEQANQLG-RLRYAV
MMAR_4307|M.marinum_M LDIR---GEDVVADLGDRARFAAADVTDEAAVASALDLAETMG-TLRIVV
MUL_0987|M.ulcerans_Agy99 LDIR---GEDVVADLGDRARFAAADVTDEAAVASALDLAETMG-TLRIVV
Mb1176|M.bovis_AF2122/97 VDLR---GDDVVGGLGDRARFAQADVTDEAAVSNALELADSLG-PVRVVV
Rv1144|M.tuberculosis_H37Rv VDLR---GDDVVGGLGDRARFAQADVTDEAAVSNALELADSLG-PVRVVV
Mflv_2127|M.gilvum_PYR-GCK IDLK---GEDVVAELGPRAKFVEANVTDPEQVAAALDAAEEFG-PLRINV
MAB_0368|M.abscessus_ATCC_1997 ADVQDEKGEALAKELGG--AFVHTDVTSEADGIAAVDAAKELG-PVRVLI
MLBr_01807|M.leprae_Br4923 THRAS--------VVPDWFFGVECDVTDNDAIGAAFKAVEEHQGPVEVLV
. : . :.*. *. .. :. :
MSMEG_4871|M.smegmatis_MC2_155 DTHGGPASGGRLVGKDGSPLD---LDGFKKTIEFYLTAVFNVMRLTAAAI
TH_3346|M.thermoresistible__bu DTHGGPAGGGRLVGKDGSPLG---LEAFETTIKYYLTAVFNIMRLTAAAI
Mvan_4251|M.vanbaalenii_PYR-1 DTHGGPAGGGRLVGKDGSPLS---MEAFETTIKFYLTAVFNIMRLSAAAI
MAV_2552|M.avium_104 VAHGGFGVAQRIVQRDGSPAD---MAAFTKTINLYLNGTYNLTRLVAAAV
MMAR_4307|M.marinum_M NCAG-TGNAIRVLSRDGVFP----LAAFRKIVDINLVGSFNVLRLAAERI
MUL_0987|M.ulcerans_Agy99 NCAG-TGNAIRVLSRDGVFS----LAAFRKIVDINLVGSFNVLRLAAERI
Mb1176|M.bovis_AF2122/97 NCAG-TGNAIRVLSRDGVFP----LAAFRKIVDINLVGTFNVLRLGAERI
Rv1144|M.tuberculosis_H37Rv NCAG-TGNAIRVLSRDGVFP----LAAFRKIVDINLVGTFNVLRLGAERI
Mflv_2127|M.gilvum_PYR-GCK NCAG-IGNAIKTLSKDGPFP----LDGFTKVIQVNLIGTFNVLRLAAERM
MAB_0368|M.abscessus_ATCC_1997 NCAG-VGWPGRTIGKDGQYASAHSLDIFSKVIGINLIGSFNLIRLAATAI
MLBr_01807|M.leprae_Br4923 ANAGITSDMLIMRMSE---------EQFQKVIDVNLTGVFRVAKLASRNM
* . : * . : * . :.: :* : :
MSMEG_4871|M.smegmatis_MC2_155 ARTEPLE--EGGRGVIVNTASIAGYEGQIGQLPYAAAKGGVLGMTLVAAR
TH_3346|M.thermoresistible__bu GRTEPLE--EGGRGVIVNTASIAGYEGQIGQLPYAAAKGGVIGMTLNAAR
Mvan_4251|M.vanbaalenii_PYR-1 AKTEPLE--EGGRGVIVNTASVAGYEGQIGQLPYSAAKGGVIGMTLVAAR
MAV_2552|M.avium_104 AAAEPRE--DGERGAIVMTASVAGYEGQIGQTAYAAAKAGVIGLTIAAAR
MMAR_4307|M.marinum_M AKTEPVGPNAEERGVIINTASVAAFDGQIGQAAYSASKGGVVGMTLPIAR
MUL_0987|M.ulcerans_Agy99 AKTEPVGPNAEERGVIINTASVAAFDGQIGQAAYSASKGGVVGMTLPIAR
Mb1176|M.bovis_AF2122/97 AKTEPIG---EERGVIINTASVAAFDGQIGQAAYSASKGGVVGMTLPIAR
Rv1144|M.tuberculosis_H37Rv AKTEPIG---EERGVIINTASVAAFDGQIGQAAYSASKGGVVGMTLPIAR
Mflv_2127|M.gilvum_PYR-GCK AKQEPIN---GERGVIINTASVAAFEGQIGQAAYSASKGGVVGMTLPIAR
MAB_0368|M.abscessus_ATCC_1997 SQEEPVD-EFGERGAIVNTASVAAFDGQIGQAAYSASKGGIVGMTLPIAR
MLBr_01807|M.leprae_Br4923 IKQ--------RFGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLAR
* : .*:.. .* ** *:*:*.*::*:: **
MSMEG_4871|M.smegmatis_MC2_155 DLSPLGIRVVTIAPGTINTPAYGQ--AADQLEEYWGPQVPFPKRMGRSTE
TH_3346|M.thermoresistible__bu DLSPLGIRVVTIAPGTINTPAYGQ--HADQLEAYWGPQVPFPKRMGRSTE
Mvan_4251|M.vanbaalenii_PYR-1 DLSPLGIRVVTIAPGTINTPAYGK--AADQLEQYWGPQVPFPKRMGRSTE
MAV_2552|M.avium_104 DLSSVGIRVNTIAPGTMKTPIMES--VGEEAIAKFAANIPFPKRLGTPAE
MMAR_4307|M.marinum_M DLASHRIRVMTIAPGLFDTPLLAS--LPEEARASLGKQVPHPSRLGSPDE
MUL_0987|M.ulcerans_Agy99 DLASHRIRVMTIAPGLFDTPLLAS--LPEEARASLGKQVPHPSRLGNPDE
Mb1176|M.bovis_AF2122/97 DLASKLIRVVTIAPGLFDTPLLAS--LPAEAKASLGQQVPHPSRLGNPDE
Rv1144|M.tuberculosis_H37Rv DLASKLIRVVTIAPGLFDTPLLAS--LPAEAKASLGQQVPHPSRLGNPDE
Mflv_2127|M.gilvum_PYR-GCK DLSREFIRVCTIAPGLFKTPLLGS--LPEEAQASLGKQVPHPARLGDPDE
MAB_0368|M.abscessus_ATCC_1997 DLSVVGIRVNTIAPGLIDTPIYGEGESAQQFKDRLAPNVLYPQRLGNPEE
MLBr_01807|M.leprae_Br4923 ELGSRSITANVVAPGFIDTEMTRA---MDSKYQEAALQAIPLGRIGAVEE
:*. * . .:*** :.* . . : *:* *
MSMEG_4871|M.smegmatis_MC2_155 YAQLAQSIVEN--DYLNGEVIRLDGALRFPPK
TH_3346|M.thermoresistible__bu YAQLAQSIIEN--DYLNGEIIRLDGALRFPPK
Mvan_4251|M.vanbaalenii_PYR-1 YAQLAQSVIEN--DYLNGEVIRLDGALRFPPK
MAV_2552|M.avium_104 YADAATFLLSN--GYVNGEVMRLDGAQRFTPK
MMAR_4307|M.marinum_M YGALAVHIIEN--PMLNGEVIRLDGAIRMAPR
MUL_0987|M.ulcerans_Agy99 YGALAVHIIEN--PMLNGEVIRLDGAIRMAPR
Mb1176|M.bovis_AF2122/97 YGALVLHIIEN--PMLNGEVIRLDGAIRMAPR
Rv1144|M.tuberculosis_H37Rv YGALVLHIIEN--PMLNGEVIRLDGAIRMAPR
Mflv_2127|M.gilvum_PYR-GCK YGALAVHIVEN--PMLNGETIRLDGAIRMAPR
MAB_0368|M.abscessus_ATCC_1997 FASLALELVTN--SYMNAETIRIDGGARLQPK
MLBr_01807|M.leprae_Br4923 IAGAVSFLASEDAGYISGAVIPVDGGLGMGH-
. . : : :.. : :**. :