For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SPVGQLISAIVLIFFGGVFAAIDAALSTVSMARVEELVREERPGAVRLARVMVERPRYINLIVLLRITCE VSATVLLAAFLDGTLGVTWGLVAAAAVMAVVSFVAIGVGPRTLGRQNAYTIALVTALPLQAISVLLLPVS RLLVLIGNALTPGRGFRNGPFASEIELREVVDLAQQRGVVADDERRMIQSVFELGDTPAREVMVPRTEMV WIEYDKTAGQATSLAVRSGHSRIPVVGENVDDIVGVVYLKDLVQRTYYSSNQGRDTSVSDVMRKPTFVPD SKPLDALLRDMQRDRVHMVLLVDEYGAIAGLVTIEDVLEEIVGEIADEYDTDEVAPVEDLGDQQYRVSAR LSIEDLGELYGIDFEEDLDVDTVGGLVALELGRVPLPGAEVTWDGLRLRAEGGPDPRGRVRIGTVLVSPV EPAPDTAEGHE*
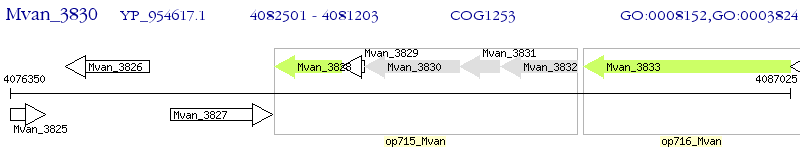
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3830 | - | - | 100% (432) | CBS domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1422 | - | 7e-34 | 30.41% (411) | hypothetical protein Mvan_1422 |
| M. vanbaalenii PYR-1 | Mvan_3094 | - | 5e-31 | 28.16% (412) | hypothetical protein Mvan_3094 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2387c | - | 0.0 | 75.88% (427) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2708 | - | 0.0 | 88.43% (432) | CBS domain-containing protein |
| M. tuberculosis H37Rv | Rv2366c | - | 0.0 | 75.88% (427) | transmembrane protein |
| M. leprae Br4923 | MLBr_00387 | guaB2 | 7e-05 | 25.22% (115) | inosine 5'-monophosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1670 | - | 0.0 | 75.46% (432) | hypothetical protein MAB_1670 |
| M. marinum M | MMAR_3676 | - | 0.0 | 77.01% (422) | hypothetical protein MMAR_3676 |
| M. avium 104 | MAV_2028 | - | 0.0 | 76.76% (426) | CBS domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_4495 | - | 0.0 | 82.25% (417) | CBS domain-containing protein |
| M. thermoresistible (build 8) | TH_1056 | - | 0.0 | 84.45% (431) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2387c|M.bovis_AF2122/97 -MTGYYQLLG-----SIVLIGLGGLFAAIDAAIS-----------TVSPA
Rv2366c|M.tuberculosis_H37Rv -MTGYYQLLG-----SIVLIGLGGLFAAIDAAIS-----------TVSPA
MMAR_3676|M.marinum_M -MKGLPQLLG-----AIALVGLGGLFAAIDAAIS-----------TVSLA
MAV_2028|M.avium_104 -MTGWSELLG-----AVALIALGGLFAAIDAAIS-----------TVSLA
Mvan_3830|M.vanbaalenii_PYR-1 -MSPVGQLIS-----AIVLIFFGGVFAAIDAALS-----------TVSMA
Mflv_2708|M.gilvum_PYR-GCK -MTNVGQLIS-----AIVLVAFGGLFAATDAALS-----------TVSIA
TH_1056|M.thermoresistible__bu -VSGLAQLVA-----AVVLVALAGVLAAVDSALN-----------TVSTA
MSMEG_4495|M.smegmatis_MC2_155 -MTGLLPLIG-----VITLVVFGGLFAAIDAALS-----------TVSIA
MAB_1670|M.abscessus_ATCC_1997 -MSSIGLLLA-----AIALVGLGGAFAAIDAAIN-----------TVSRA
MLBr_00387|M.leprae_Br4923 MIRGMSNLKESSDFVASSYVRLGGLMDDPAATGGDNPHKVAMLGLTFDDV
: * : :.* : :: . *.. .
Mb2387c|M.bovis_AF2122/97 RVDELVRDQRPGAGSLRKVMADRPRYVNLVVLLRTSCEITATALLVVFIR
Rv2366c|M.tuberculosis_H37Rv RVDELVRDQRPGAGSLRKVMADRPRYVNLVVLLRTSCEITATALLVVFIR
MMAR_3676|M.marinum_M RVQELVREARPGAVALSKVMEDRPRYINLVVLLRIVCEITATALLVVFFR
MAV_2028|M.avium_104 RVQELVRDERPGAVALSKVMSERPRYINLVVLLRITCEITATVLLVVFLY
Mvan_3830|M.vanbaalenii_PYR-1 RVEELVREERPGAVRLARVMVERPRYINLIVLLRITCEVSATVLLAAFLD
Mflv_2708|M.gilvum_PYR-GCK RVEELVREERPGAVRLLRLMVDRPRYINLIVLLRMTCEVSATVLMAAYLD
TH_1056|M.thermoresistible__bu RVGELVRDERPGAVRLARVVADRPKYVNLIVLLRIACEVSATVLLVAFLD
MSMEG_4495|M.smegmatis_MC2_155 RIDELVRDERPGAARLATVVAERPRYINLVVLLRIICEVTATVLLASILD
MAB_1670|M.abscessus_ATCC_1997 RVDELVREERPGAVRLSAVIRERPRYVNLVVLLRTTCEILSTVFLVGYLT
MLBr_00387|M.leprae_Br4923 LLLPAASDVVPATADISSQLTKKIRLKVPLVSSAMDTVTEARMAIAMARA
: . : *.: : : .: : :* : :.
Mb2387c|M.bovis_AF2122/97 YHFSMVWG---LYLAAGIMVLASFVVVGVGPRTLGRQNAYSISLATALPL
Rv2366c|M.tuberculosis_H37Rv YHFSMVWG---LYLAAGIMVLASFVVVGVGPRTLGRQNAYSISLATALPL
MMAR_3676|M.marinum_M HNLSMNGG---LFLAAAIMVVTSFVVIGVGPRTLGRQNAYSISLGTAVPL
MAV_2028|M.avium_104 DNFGLRWG---LFGAAAIMVVTSFVVMGVGPRTLGRQNAYSIALTTVLPL
Mvan_3830|M.vanbaalenii_PYR-1 GTLGVTWG---LVAAAAVMAVVSFVAIGVGPRTLGRQNAYTIALVTALPL
Mflv_2708|M.gilvum_PYR-GCK DYLGVSRG---LMAAAAIMTVVSFVVVGVGPRTLGRQNAYTIALVTALPL
TH_1056|M.thermoresistible__bu GYLGLRWG---MVTAAAVMVVVSFVAIGVGPRTVGRQNAYSIALMAALPL
MSMEG_4495|M.smegmatis_MC2_155 GLLGVSWG---LVTAAAIMVVTSFVAMGVGPRTVGRQNAYSIALMSAIPL
MAB_1670|M.abscessus_ATCC_1997 ARISLGAA---LTISAIVMVVTSFVAIGVGPRTLGRQHAYSIALGAAVPL
MLBr_00387|M.leprae_Br4923 GGMGVLHRNLPVGEQAGQVETVKRSEAGMVTDPVTCRPDNTLAQVGALCA
:.: : * : .. *: . .: : ::: .:
Mb2387c|M.bovis_AF2122/97 RL--------------------------------ISWLLMPISRLLVLLG
Rv2366c|M.tuberculosis_H37Rv RL--------------------------------ISWLLMPISRLLVLLG
MMAR_3676|M.marinum_M QV--------------------------------ISWLLMPISRLLVLLG
MAV_2028|M.avium_104 QV--------------------------------ISWLLMPISRLLVVLG
Mvan_3830|M.vanbaalenii_PYR-1 QA--------------------------------ISVLLLPVSRLLVLIG
Mflv_2708|M.gilvum_PYR-GCK QA--------------------------------ISVVLTPISRLLVLIG
TH_1056|M.thermoresistible__bu QT--------------------------------ISVLLTPISRLLVLLG
MSMEG_4495|M.smegmatis_MC2_155 QA--------------------------------LSVLLTPISRLLVLLG
MAB_1670|M.abscessus_ATCC_1997 QV--------------------------------ISVLLGPISRLLILLG
MLBr_00387|M.leprae_Br4923 RFRISGLPVVDDSGALAGIITNRDMRFEVDQSKQVAEVMTKTPLITAAEG
: :: :: . : *
Mb2387c|M.bovis_AF2122/97 NALTPGRGFR-------------NGPFASEIELREVVDLAQQRGVVAADE
Rv2366c|M.tuberculosis_H37Rv NALTPGRGFR-------------NGPFASEIELREVVDLAQQRGVVAADE
MMAR_3676|M.marinum_M NALTPGRGFR-------------NGPFASEVELREVVDLAQQRGVVAADE
MAV_2028|M.avium_104 NAVTPGRGLR-------------NGPFASEIELREVVDLAQQRGVVAAEE
Mvan_3830|M.vanbaalenii_PYR-1 NALTPGRGFR-------------NGPFASEIELREVVDLAQQRGVVADDE
Mflv_2708|M.gilvum_PYR-GCK NALTPGRGFR-------------NGPFASEIELREVVDLAQQRGVVADDE
TH_1056|M.thermoresistible__bu NALTPGRGFR-------------NGPFASEIELREVVDLAQQRGVVADDE
MSMEG_4495|M.smegmatis_MC2_155 NALTPGRGFR-------------NGPFASEIELREVVDLAQQRGVVADEE
MAB_1670|M.abscessus_ATCC_1997 NAVTPGRGFR-------------NGPFASEIELREVVDMAQQRGVVADEE
MLBr_00387|M.leprae_Br4923 VSADAALGLLRRNKIEKLPVVDGHGRLTGLITVKDFVKTEQHPLATKDND
: .. *: :* ::. : :::.*. *: .. ::
Mb2387c|M.bovis_AF2122/97 -RRMIESVFELGDTPAREVMVPRTEMIWIESDKTAGQAMTLAVRSGHSRI
Rv2366c|M.tuberculosis_H37Rv -RRMIESVFELGDTPAREVMVPRTEMIWIESDKTAGQAMTLAVRSGHSRI
MMAR_3676|M.marinum_M -RRMIESVFELGDTPAREVMVPRTEMVWIEGDKSPSQAMTLAVRSGHSRI
MAV_2028|M.avium_104 -RRMIQSVFELGDTPAREVMVPRTEMIWIESDKSASQAINLAVRSGHSRI
Mvan_3830|M.vanbaalenii_PYR-1 -RRMIQSVFELGDTPAREVMVPRTEMVWIEYDKTAGQATSLAVRSGHSRI
Mflv_2708|M.gilvum_PYR-GCK -RRMIQSVFELGDTAAREVMVPRTEMVWIESDKTAGQATSLAVRSGHSRI
TH_1056|M.thermoresistible__bu -RRMIQSVFELGDTPAREVMVPRTEMVWIEQDKTAGQATSLAVRSGHSRI
MSMEG_4495|M.smegmatis_MC2_155 -RRMIQSVFELGDTAAREVMVPRTEMVWIEADKTAGQATSLAVRSGHSRI
MAB_1670|M.abscessus_ATCC_1997 -RRMIQSVFELGDTPAREVMVPRTEMVWIESDKTAGQATSLAVRSGHSRI
MLBr_00387|M.leprae_Br4923 GRLLVGAAVGVGGDAWVRAMMLVDAGVDVLIVDTAHAHNRLVLDMVGKLK
* :: :.. :*. . ..*: : : .:. *.: .
Mb2387c|M.bovis_AF2122/97 PVIGENVDDIVGVVYLK----DLVEQTFCSTNGGRETTVARVMRPAVFVP
Rv2366c|M.tuberculosis_H37Rv PVIGENVDDIVGVVYLK----DLVEQTFCSTNGGRETTVARVMRPAVFVP
MMAR_3676|M.marinum_M PVVGENVDDILGVVYLK----DLVRQTFCAPDQGRKIDVSDLMRPAVFVP
MAV_2028|M.avium_104 PVIGENVDDIVGVVYLK----DLVQQALLPGGSGRATPVSQVMRPAVFVP
Mvan_3830|M.vanbaalenii_PYR-1 PVVGENVDDIVGVVYLK----DLVQRTYYSSNQGRDTSVSDVMRKPTFVP
Mflv_2708|M.gilvum_PYR-GCK PVIGENVDDVVGVVYLK----DLVQKTYYSNNGGRDTAVSAVMRKPVFVP
TH_1056|M.thermoresistible__bu PVIGENVDDIVGVVYLK----DLVQRTYYSTDGGRGTLVSEVMRPAVFVP
MSMEG_4495|M.smegmatis_MC2_155 PVIGENVDDIVGVVYLK----DLVQQTYYSTNGGRDTTVAQVMRPAVFVP
MAB_1670|M.abscessus_ATCC_1997 PVVGENVDDVVGVVFLK----DLVQQTYYSVNGGRDVTVAQVMRPAVFVP
MLBr_00387|M.leprae_Br4923 VEIGDRVQVIGGNVATRSAAAALVEAGADAVKVGVGPGSTCTTRVVAGVG
:*:.*: : * * : **. . * : * . *
Mb2387c|M.bovis_AF2122/97 DSKPLDALLREMQRDRNHMALLVDEYGAIAGLVSIEDVLEEIVGEIADEY
Rv2366c|M.tuberculosis_H37Rv DSKPLDALLREMQRDRNHMALLVDEYGAIAGLVSIEDVLEEIVGEIADEY
MMAR_3676|M.marinum_M DSKPLDALLREMQHDRNHMALLVDEYGAIAGLVSIEDVLEEIVGEIADEY
MAV_2028|M.avium_104 DSKPLDALLREMQRDRNHMALLVDEYGAIAGLVSIEDVLEEIVGEIADEY
Mvan_3830|M.vanbaalenii_PYR-1 DSKPLDALLRDMQRDRVHMVLLVDEYGAIAGLVTIEDVLEEIVGEIADEY
Mflv_2708|M.gilvum_PYR-GCK DSKPLDALLSEMQRDRVHMVLLVDEYGAIAGLVTIEDVLEEIVGEIADEY
TH_1056|M.thermoresistible__bu DSKPLDELLREMQRDRNHMALLVDEYGAIAGLVTIEDVLEEIVGEIADEY
MSMEG_4495|M.smegmatis_MC2_155 DSKPLDELLSEMQRDRNHMALLVDEYGATAGLVTIEDVLEEIVGEIADEY
MAB_1670|M.abscessus_ATCC_1997 DSKPLDELLREMQQHRKHMALLVDEYGAIAGLVTIEDVLEEIVGEIADEY
MLBr_00387|M.leprae_Br4923 APQITAILEAVAACGPAGVPVIADGGLQYSGDIAKALAAGASTTMLGSLL
.: * : ::.* :* :: . . :..
Mb2387c|M.bovis_AF2122/97 DQAETAPVEDLGDKRFRVSARLPIEDVGELYGVEFDDDLDVDTVGGLLAL
Rv2366c|M.tuberculosis_H37Rv DQAETAPVEDLGDKRFRVSARLPIEDVGELYGVEFDDDLDVDTVGGLLAL
MMAR_3676|M.marinum_M DEAETAPVEALGDKRFRVSARLPIEDVGELYGIEFDGDLDVDTVGGLLAL
MAV_2028|M.avium_104 DQAETAPIEDLGDKRFRVSARLPIEDLGELYGVEFDDDLDVDTVGGLLAL
Mvan_3830|M.vanbaalenii_PYR-1 DTDEVAPVEDLGDQQYRVSARLSIEDLGELYGIDFEEDLDVDTVGGLVAL
Mflv_2708|M.gilvum_PYR-GCK DTDEVAPIEDLGELSYRVSARLPIEDLSELYGIDFDEGLDVDTVGGLVAL
TH_1056|M.thermoresistible__bu DTDEVAPIEDLGDNRYRVSARLPIEDLGELYGREFDGDLDVDTVGGLVAL
MSMEG_4495|M.smegmatis_MC2_155 DTDEVAPVDEIDDHIYRVSARLPIEDLAELYELELDEDLDVDTVGGLMAF
MAB_1670|M.abscessus_ATCC_1997 DHDEVAPVEELGDKMFRVSARLPIEDLGELYGIEFDEDLDVETVGGLLAL
MLBr_00387|M.leprae_Br4923 AGTAEAPGELIFVNGKQFKSYRGMGSLGAMQGRGGDKSYSKDRYFADDAL
** : : :..: : .:. : : . . : . *:
Mb2387c|M.bovis_AF2122/97 ELG---------RVPLPG-AEVISHGLRLHAEGGTDHRGRVRIGTVLLSP
Rv2366c|M.tuberculosis_H37Rv ELG---------RVPLPG-AEVISHGLRLHAEGGTDHRGRVRIGTVLLSP
MMAR_3676|M.marinum_M ELG---------RVPLPG-AEVIFHGLRLRAEGGPDHRGRVRIGTVLVSP
MAV_2028|M.avium_104 ELG---------RVPLPG-AEVISHGLRLHAEGGTDHRGRVRVGTVLLSP
Mvan_3830|M.vanbaalenii_PYR-1 ELG---------RVPLPG-AEVTWDGLRLRAEGGPDPRGRVRIGTVLVSP
Mflv_2708|M.gilvum_PYR-GCK ELG---------RVPLPG-AEVEWDGLRLRAEGGPDPRGRVRIGTVLVSP
TH_1056|M.thermoresistible__bu ELG---------RVPLPG-AEVTWDGLRLRAEGGPDHRGRVRIGTVLVSP
MSMEG_4495|M.smegmatis_MC2_155 ELG---------RVPLPG-AEVTWEGLRLRAEGGPDHRGRVRINTVLVCP
MAB_1670|M.abscessus_ATCC_1997 DLG---------RVPLPG-AKVASHGLLLQAEGGPDARGRVRIGTILVQP
MLBr_00387|M.leprae_Br4923 SEDKLVPEGIEGRVPFRGPLSSVIHQLVGGLRAAMGYTGSPTIEVLQQAQ
. . ***: * . . * ... . * : .:
Mb2387c|M.bovis_AF2122/97 AE-------PDGADDEEADHPG--------------
Rv2366c|M.tuberculosis_H37Rv AE-------PDGADDEEADHPG--------------
MMAR_3676|M.marinum_M METEE----TDGAGDGAAETGADG------------
MAV_2028|M.avium_104 VE-------SDGSPDGEEPEHHD-------------
Mvan_3830|M.vanbaalenii_PYR-1 VEPA--------PDTAEGHE----------------
Mflv_2708|M.gilvum_PYR-GCK VEPP--------NDDDEGHE----------------
TH_1056|M.thermoresistible__bu VEPED-------ADDTEDRDGADD------------
MSMEG_4495|M.smegmatis_MC2_155 TEPVEPELHTVPGDENSGRTRRGDRGDRSRGGEKDE
MAB_1670|M.abscessus_ATCC_1997 DPD---------AESDEKAEERDE------------
MLBr_00387|M.leprae_Br4923 FVRITPAGLKESHPHDVAMTVEAPNYYPR-------