For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DELVAARDQMERLVRVIVAIGADLDLDVTLRRIVTGARELSGARYGALGIRGADGTWASFVCDGMEEDAA RALGGLAAGEGIRIDDLTGYSGVAGLRSQKTPMRALLGVPITVRGADFGALYLADDQPDRVFSDSQEGAV RALATAAAAAIDNARLFERERESAKWTKASREITTALLSGDPQTGPLQLIVNLALELADAEQAILLVPGE PDLPGDEADTLVVAATAGRYASEVIGRQVPMDGSTTGGVARRGLPLITDSFQYPIEGFTDAGERSAIVMP LIADEAVLGVIAVARRPHQPPFDNDYLELVSDFARHAAIALALAAGREHALNQELAQADSVDAALHAAAE ELRRLWRARRVLAVTFPTHGDSVESAPKVVAVGEDTHWADLSAPTRLTLSALRDGDLLTPNTAQPGTAGI AMQHPEGILVVWLDLTDQRSFTLEDQTLLTVLAGRLSQGLQRVHRVDQQRETALALQHAILGPADLPHGF AVRYQAASRPLQVGGDWYDVVDLDDGRIAMIVGDCVGHGLPAATVMGQVRSACRALLFDNPSPAAALAGM DRFAARLPGAQCATAVCAVLDPATGELVYSSAGHPPPILMHADGSARLLEGAHTIALGLRVGWSRPEGRL SIPAGATLMLYTDGLVERRRTLLEDGISRAAELLEGHRTSDLEDLADQIMSGLAPSVGYQDDVALLLYRH PAPLQLNFPADANHLAPARAALRSWLTSARVEPARVMDVLVAAGEALANAIEHGHRHTPDGTVSLRATAL VDRVRLTVADTGSWKVPQPGRDSRRGRGIALMRALMHDVEIDIGRSGTSVHLDTRID*
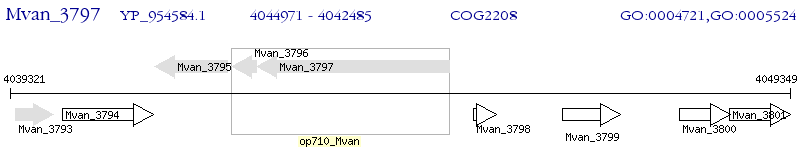
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3797 | - | - | 100% (828) | putative GAF sensor protein |
| M. vanbaalenii PYR-1 | Mvan_1931 | - | e-175 | 63.47% (501) | multi-sensor signal transduction histidine kinase |
| M. vanbaalenii PYR-1 | Mvan_1413 | - | 5e-85 | 45.69% (383) | GAF sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2052c | - | 2e-54 | 35.65% (359) | histidine kinase response regulator |
| M. gilvum PYR-GCK | Mflv_4521 | - | 0.0 | 74.58% (826) | putative GAF sensor protein |
| M. tuberculosis H37Rv | Rv2027c | - | 2e-54 | 35.65% (359) | histidine kinase response regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3890c | - | 2e-50 | 36.06% (355) | histidine kinase response regulator |
| M. marinum M | MMAR_3479 | - | 1e-101 | 56.94% (360) | two-component sensor histidine kinase |
| M. avium 104 | MAV_2108 | - | 0.0 | 74.94% (830) | stage II sporulation protein E (SpoIIE) |
| M. smegmatis MC2 155 | MSMEG_3941 | - | 7e-60 | 42.90% (338) | GAF family protein |
| M. thermoresistible (build 8) | TH_2781 | - | 3e-97 | 40.98% (510) | PUTATIVE stage II sporulation protein E (SpoIIE) |
| M. ulcerans Agy99 | MUL_3853 | rsbU | 2e-49 | 31.96% (438) | regulator of sigma subunit, anti-anti-sigma factor RsbU |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3797|M.vanbaalenii_PYR-1 ------------------------------------MDELVAARDQMERL
Mflv_4521|M.gilvum_PYR-GCK ------------MADDRNLPAFRRHGRQEGDRTDAEFDELIAARNQMDHL
MAV_2108|M.avium_104 --------------------------------MHTRLDELMAARDQLEQL
MMAR_3479|M.marinum_M --------------------------------MHAQLDELLAARDQMEQL
MSMEG_3941|M.smegmatis_MC2_155 ------------------------------MGQSHTDDSPEAVEVRMAAL
Mb2052c|M.bovis_AF2122/97 MTHPDRANVNPGSPPVRETLSQLRLRELLLEVQDRIEQIVEG-RDRLDGL
Rv2027c|M.tuberculosis_H37Rv MTHPDRANVNPGSPPLRETLSQLRLRELLLEVQDRIEQIVEG-RDRLDGL
MAB_3890c|M.abscessus_ATCC_199 MVEDVGAQPNR----VTAELAHLQLRELLAEVQGRIEQIVDGTRGRMDAL
TH_2781|M.thermoresistible__bu --------------------------------------------MSPDFG
MUL_3853|M.ulcerans_Agy99 --------------------------------MLLLDLLRQRTYSFVRSV
Mvan_3797|M.vanbaalenii_PYR-1 VRVIVAIGADLDLDVTLRRIVTGARELSGARYGALGIRGADGTWASFVCD
Mflv_4521|M.gilvum_PYR-GCK VRAIVAIGSDLDFSVTLERIVTAARELSGARYAGLGIRAAGGNLVSFVES
MAV_2108|M.avium_104 VRVIAEIGSDLDLDVTLRRVLKAAMELTGARYAVLSSRASDGGLLSFVHA
MMAR_3479|M.marinum_M LRVIVEIGSDLDLDATLRRIVLAARELTSAPYGALAVRDPEGTLIRFIHA
MSMEG_3941|M.smegmatis_MC2_155 LQDMASLDAGLEPESTLHGILDAIMRVSGACYGAAGLCAPDGTPATFLQA
Mb2052c|M.bovis_AF2122/97 IDAILAITSGLKLDATLRAIVHTAAELVDARYGALGVRGYDHRLVEFVYE
Rv2027c|M.tuberculosis_H37Rv IDAILAITSGLKLDATLRAIVHTAAELVDARYGALGVRGYDHRLVEFVYE
MAB_3890c|M.abscessus_ATCC_199 LDAVMAVSSGLELDATLREIVCAASELVSARYGALGVVGSDEKLAEFVYE
TH_2781|M.thermoresistible__bu ARYAAALAAYLE-DSSERRLAVAHDLGRQALLGRISVLQIVEDHVRLVDE
MUL_3853|M.ulcerans_Agy99 HRRDGYMAAEMDWDKNVGTANTVRRIFENVPTMLVGLEGPEHRFVAANAA
: : :. . . . .
Mvan_3797|M.vanbaalenii_PYR-1 GMEEDAARALGGLAAGEGIR-----------IDDLTGYSGVAGLRSQKTP
Mflv_4521|M.gilvum_PYR-GCK GDEFDVA-ALAGLPIDGGLR-----------IDDLRDHPEITDLQALNRP
MAV_2108|M.avium_104 GLDADTARQLGELAVGDGLR-----------IDDVSVDPRSAHLSAHDPP
MMAR_3479|M.marinum_M GMDDDVARRIGHLPVGKGVLGVSLVETPALRLDDLTMHPAACGFPEHHPP
MSMEG_3941|M.smegmatis_MC2_155 G---ALAQAVPDSPD-RDMLGFLRNRTGPLRIDGLARSAAEVGLPELLPP
Mb2052c|M.bovis_AF2122/97 GIDEETRHLIGSLPEGRGVLGALIEEPKPIRLDDISRHPASVGFPLHHPP
Rv2027c|M.tuberculosis_H37Rv GIDEETRHLIGSLPEGRGVLGALIEEPKPIRLDDISRHPASVGFPLHHPP
MAB_3890c|M.abscessus_ATCC_199 GIDEATRDLIGPLPTGHGVLGVVIDEAKPLRLRDIAAHPASVGFPANHPP
TH_2781|M.thermoresistible__bu LSRRQPVDTDTALQFLLQAL--------------AAVDVAVRGYLEGTRR
MUL_3853|M.ulcerans_Agy99 YRRFSPTFTAVGQPAREVYPELE---------SQQIYHMLDRVYETGEPQ
Mvan_3797|M.vanbaalenii_PYR-1 MRALLGVPITVRGADFGALYLADDQPDRVFSDSQEGAVRALATAAAAAID
Mflv_4521|M.gilvum_PYR-GCK IRALLAIPVTVRGADFGSLYLADDRPGRVFSDAQESAVRALATAAAAAID
MAV_2108|M.avium_104 LRALLGIPITVRAANFGNLYLADDRQGRVFTDSQEGAVRAMATAAAAAID
MMAR_3479|M.marinum_M MRAFLAVPITIRGTVFGSLYLTHVDPGRKFSESDEFAARALAFAAGVAID
MSMEG_3941|M.smegmatis_MC2_155 MRAVLGVPLVSHGEVLGSLYVADSRAGFEFSDAHVGVVEVLATVAADVVH
Mb2052c|M.bovis_AF2122/97 MRTFLGVPVRIRDEVFGNLYLTEKADGQPFSDDDEVLVQALAAAAGIAVD
Rv2027c|M.tuberculosis_H37Rv MRTFLGVPVRIRDEVFGNLYLTEKADGQPFSDDDEVLVQALAAAAGIAVD
MAB_3890c|M.abscessus_ATCC_199 MRTFLGVPVQVRGEVFGRLYLTEKLDGQEFTEDDEVVVRALAGAAGIAIE
TH_2781|M.thermoresistible__bu YEQQRARAEGLAGRDTFRSALVNAMQEAFFVMDRTGSVIDINEAFAEITG
MUL_3853|M.ulcerans_Agy99 SGAEWRLQADFDGSGIDERYFDFLVTPRRREDGSIEGVQILFDDVTTRVR
. . :
Mvan_3797|M.vanbaalenii_PYR-1 NARLFERERESAKWTKASREITTALLSGDPQ-TGPLQ-LIVNLALELADA
Mflv_4521|M.gilvum_PYR-GCK NARLFEREREAARWTKASREITTALLSGDPQ-TGPLQ-LIVNRALELAGA
MAV_2108|M.avium_104 NARLFERERESAKWTKASREITTALLSGDPQ-TGPLQ-LIVNRALELAGA
MMAR_3479|M.marinum_M NAQVFERERTAVKWMEASREITTALMSSTESRREPLQ-LIAQRACTLTDA
MSMEG_3941|M.smegmatis_MC2_155 SVWHLAKVRTVARWVSASREITMALLGMSGGEDAPLR-LIVGRAAELTVA
Mb2052c|M.bovis_AF2122/97 NARLFEESRTREAWIEATRDIGTQMLAGADP-AMVFR-LIAEEALTLMAG
Rv2027c|M.tuberculosis_H37Rv NARLFEESRTREAWIEATRDIGTQMLAGADP-AMVFR-LIAEEALTLMAG
MAB_3890c|M.abscessus_ATCC_199 NARLYQHARRREQWQRATADITAELLGGVDP-GKALH-LVASSAALLAEA
TH_2781|M.thermoresistible__bu YDGHDLPYRWPHPWVVDEPRADEDLLELLQHGRVQLERQIRHRSGHLAWV
MUL_3853|M.ulcerans_Agy99 --ARLAAEARVEELSERYRHLRDSAIVMQQALLAPSVPVVSGPDIAAQYL
: :
Mvan_3797|M.vanbaalenii_PYR-1 EQAILLVPGEPDLPGDEADTLVVAATAGRYASEVIGRQVPMDGSTTGGVA
Mflv_4521|M.gilvum_PYR-GCK EQAILLVPLEPDVPAEDVDTLIVAATAGRYSSEVIGRQVPMAGSTTGGVA
MAV_2108|M.avium_104 EQAILLVPREPESPGREADTLVVSATAGRYASQVIGRQVPMDGSTTGGVA
MMAR_3479|M.marinum_M EQAIVLVPVESDLATDEIDTLVVSAAVGLHAEEVVGQQVPVRGSTCGDVF
MSMEG_3941|M.smegmatis_MC2_155 SQAAILVPSEADRAPEEVRSLTVAVAAGARADEVEGQQVPVSGSTSGRVF
Mb2052c|M.bovis_AF2122/97 AATLVAVPLDDEAPACEVDDLVIVEVAGEISPAVKQMTVAVSGTSIGGVF
Rv2027c|M.tuberculosis_H37Rv AATLVAVPLDDEAPACEVDDLVIVEVAGEISPAVKQMTVAVSGTSIGGVF
MAB_3890c|M.abscessus_ATCC_199 DFAFIALPAGDEE---DTAELVVAVCEGKGAEVLNDKLIPIDGSTSGSTF
TH_2781|M.thermoresistible__bu AISVNVVNPNGTAFVGTMRDITAAHKAAARESALTRLATAAGVATS----
MUL_3853|M.ulcerans_Agy99 VAAQDTAAGGDWFDAVALGDRLVLIVGDVVGHGVEAAAVMSQLRTALRLQ
: : :
Mvan_3797|M.vanbaalenii_PYR-1 RRGLPLITDSFQYPIEGFTDAGERSAIVMPLIADEAVLGVIAVARRPHQP
Mflv_4521|M.gilvum_PYR-GCK RRGLPMITDSFQYPIEGFTDVGERSAIVMPLIVDDAVLGVIAVARRPNQP
MAV_2108|M.avium_104 RRGLPLITDSFQYPIEGFTDVGERSAIVIPLIADGAVLGVIAVARDPQQP
MMAR_3479|M.marinum_M RSGEPLISEAFSYPIEAFTDVAQRSAIVMPLSAHEKVAGVIAVARGADQP
MSMEG_3941|M.smegmatis_MC2_155 RTGESTMTDGFRYPITSFTDQGRRPAIVVPLRSATNNLGVIAVARDADDP
Mb2052c|M.bovis_AF2122/97 HDRTPRRFDRLDLAVDG--PVEPGPALVLPLRAADTVAGVLVALRSADEQ
Rv2027c|M.tuberculosis_H37Rv HDRTPRRFDRLDLAVDG--PVEPGPALVLPLRAADTVAGVLVALRSADEQ
MAB_3890c|M.abscessus_ATCC_199 IDHEPRNVGRLAVNLAEGTDLAFGPALVLPLGGGESTAGVLILLRTVGAI
TH_2781|M.thermoresistible__bu ------VSEVLSIMLDECRPAIDSRRVIAVTWAREDAEPTIQVVGDPPAS
MUL_3853|M.ulcerans_Agy99 IAAGRSIGEALEALDAFHEHVPGSNSATLCVGSLDFTTGEFQYCTAGHPP
: :
Mvan_3797|M.vanbaalenii_PYR-1 PFDNDYLELVSDFARHAAIALALAAGREHALNQELAQADSVDAALHAAAE
Mflv_4521|M.gilvum_PYR-GCK AFDEDYLELVSDFARHASIALALAAGREHALNRQLAQADSVSEAMAGAAE
MAV_2108|M.avium_104 PFGNDYLDLVSDFARHAAIALALAAGRQHALNQELAQADTVDEAVRAAAE
MMAR_3479|M.marinum_M PFDARCLDLVSQFATHAAIALLLASGREDAR-QLTILAERERIAHDLHDH
MSMEG_3941|M.smegmatis_MC2_155 PFDVDEMAIMEDFAHHAAVALTLCRANEQAR-QLTVLSDRERIARDLHDH
Mb2052c|M.bovis_AF2122/97 PFSDKQLDMMAAFADQAALAWRLATAQRQMR-EVEILTDRDRIARDLHDH
Rv2027c|M.tuberculosis_H37Rv PFSDKQLDMMAAFADQAALAWRLATAQRQMR-EVEILTDRDRIARDLHDH
MAB_3890c|M.abscessus_ATCC_199 AFDEEQLDMAASFAGQAALALEQAEVRLAKH-ELDVLADRDRIARDLHDH
TH_2781|M.thermoresistible__bu TWADLDESLRTTLLAARDWPPLTVQPVSDAG-----QNTSRGIVAVLSGT
MUL_3853|M.ulcerans_Agy99 PLVVTAEAKARYLEPSGAGPLGSDTGFPVRT----ELIGVGDVVLLYTDG
. : . .
Mvan_3797|M.vanbaalenii_PYR-1 ELRRLWRARRVLAVTFPTHGDSVES---APKVVAVGEDTHWADLSAPTRL
Mflv_4521|M.gilvum_PYR-GCK ELRRLWRARRVLAVTFPTRSEAADV---EPVVVSAGEHTTWRELPDGIRQ
MAV_2108|M.avium_104 ELRRLWRARRVLAVTFPSHTSATETAFGPPEVVSVGEPTQWADLPSHMHQ
MMAR_3479|M.marinum_M VIQRLFASGMDLQGTLA---------------------------------
MSMEG_3941|M.smegmatis_MC2_155 VIQRLFLAGMDLQGTIA---------------------------------
Mb2052c|M.bovis_AF2122/97 VIQRLFAVGLTLQGAAP---------------------------------
Rv2027c|M.tuberculosis_H37Rv VIQRLFAVGLTLQGAAP---------------------------------
MAB_3890c|M.abscessus_ATCC_199 VIQRLFAVGLAMQSTHR---------------------------------
TH_2781|M.thermoresistible__bu PATVLWLEHRVPRPVGID--------------------------------
MUL_3853|M.ulcerans_Agy99 LIERPGRPLGASTAEFAD--------------------------------
Mvan_3797|M.vanbaalenii_PYR-1 TLSALRDGDLLTPNTAQPGTAGIAMQHPEGILVVWLDLTDQRSFTLEDQT
Mflv_4521|M.gilvum_PYR-GCK ILLSMRRGDLLTPNAAEPGTAGIAVQHPEGILVVWIELAEHRSFTLEDQT
MAV_2108|M.avium_104 ALGSLRDGDLLTPNTTQPGTAGIALQHPDGVLVVWIDLTDERSFTLEDQT
MMAR_3479|M.marinum_M ------------------------------------------------RA
MSMEG_3941|M.smegmatis_MC2_155 ------------------------------------------------RT
Mb2052c|M.bovis_AF2122/97 ------------------------------------------------RA
Rv2027c|M.tuberculosis_H37Rv ------------------------------------------------RA
MAB_3890c|M.abscessus_ATCC_199 ------------------------------------------------RS
TH_2781|M.thermoresistible__bu -----------------------------------------------GRR
MUL_3853|M.ulcerans_Agy99 -------------------------------------------------L
Mvan_3797|M.vanbaalenii_PYR-1 LLTVLAGRLSQGLQRVHRVDQQRETALALQHAILGPADLPHGFAVRYQAA
Mflv_4521|M.gilvum_PYR-GCK LLTVLAGRLSAGLQRVHQVDQQRETALALQHAILGPAHLPPGFTVRYQAA
MAV_2108|M.avium_104 LLTVLAGRLGQGLQRVHQVDQQRETALALQHAILGPADLPNGFAVRYQAA
MMAR_3479|M.marinum_M RSPEVIDRLNRTVDDLQTIIEEIRSTIFQLKSPS---GRDGGFRQRIQQV
MSMEG_3941|M.smegmatis_MC2_155 KSDLLKERLSRTVDDLQSIIDEIRTAIFDLQSPA---ASAMSFRQRIQAA
Mb2052c|M.bovis_AF2122/97 RVPAVRESIYSSIDDLQEIIQEIRSAIFDLHAGP---SRATGLRHRLDKV
Rv2027c|M.tuberculosis_H37Rv RVPAVRESIYSSIDDLQEIIQEIRSAIFDLHAGP---SRATGLRHRLDKV
MAB_3890c|M.abscessus_ATCC_199 VDPTVAARLADHIDQLHEVIQEIRTAIFDLHTN------DESLRATLRST
TH_2781|M.thermoresistible__bu LINALVGHVSLAVQHVRRFEIARESSLTLQRAMLPDIVPPPGFAVRYEPA
MUL_3853|M.ulcerans_Agy99 AANITSGGGFVIDAQSRPIDRLCSETLELLLRSTGYNDDITLLAVQRRAP
: . :: :
Mvan_3797|M.vanbaalenii_PYR-1 SRPLQVGGDWYDVVDLDDGRIAMIVGDCVGHGLPAATVMGQVRSACRALL
Mflv_4521|M.gilvum_PYR-GCK SRPLQVGGDWYDVVDLDDGRIALIVGDCVGHGLAAATVMGQVRSACRALL
MAV_2108|M.avium_104 TRPLQVGGDWFDIVDLEDGRVALVVGDCVGHGLASATVMGQVRSACRALL
MMAR_3479|M.marinum_M VADLTEHRDVVTTIHMHG---PMSAVQGELADHAEAVATEAISNAVRHSG
MSMEG_3941|M.smegmatis_MC2_155 VAAVTDNSPLATTVRISG---PIVAVDPALADDAEAVIVEAISNAVRHSG
Mb2052c|M.bovis_AF2122/97 IDQLAIP-ALHTTVQYTG---PLSVVDTVLANHAEAVLREAVSNAVRHAN
Rv2027c|M.tuberculosis_H37Rv IDQLAIP-ALHTTVQYTG---PLSVVDTVLANHAEAVLREAVSNAVRHAN
MAB_3890c|M.abscessus_ATCC_199 INELTAETGLRVSVRMSG---PLDVVPASTSGDAEAVVREAVSNVVRHAR
TH_2781|M.thermoresistible__bu VRPLEIGGDWYDVLPLDDHQIGIIVGDCVGRGLPAAAVMGQLRSSARALL
MUL_3853|M.ulcerans_Agy99 TPPLNITEDATIHAARAIRSRLRQWLSEIGADSGDISDIVHAISEFVENA
: . .
Mvan_3797|M.vanbaalenii_PYR-1 FDNPSPAAALAGMDRFAARLPGAQCATAVCAVLDPATGELVYSSAGHPPP
Mflv_4521|M.gilvum_PYR-GCK LENPSPGAALSGMDRFAARLPGAQCTTAVVMVLDPASGEIAYSSAGHPPP
MAV_2108|M.avium_104 IENPSPAAALAGLDRFAARLPGAQCTTAVCAVLTPDTGELVYSSAGHPPP
MMAR_3479|M.marinum_M AGRVTLDVSVGDMFSLEVVDDGCGISPDVSRRSGLANMRSRAEKLGGTCG
MSMEG_3941|M.smegmatis_MC2_155 ATSVTISIEVSDRLDIEVVDNGRGIPANTARCSGLANLKARAQQAGGSCQ
Mb2052c|M.bovis_AF2122/97 ATSLAINVSVEDDVRVEVVDDGVGISGDITE-SGLRNLRQRADDAGGEFT
Rv2027c|M.tuberculosis_H37Rv ATSLAINVSVEDDVRVEVVDDGVGISGDITE-SGLRNLRQRADDAGGEFT
MAB_3890c|M.abscessus_ATCC_199 ARELSVTVSVDDELVIEVVDDGIGIPDVVAR-SGLSGLANRAAQSGGTFS
TH_2781|M.thermoresistible__bu LSGAEPARLLENLDAVAALIPDAYCATVLLAVLDTESGTLRYSNAGHVPA
MUL_3853|M.ulcerans_Agy99 VEHGYSTDVSDGLAVAAALGGDGKLRVAVIDHGTWKHHREGERGRGRGLA
. . . *
Mvan_3797|M.vanbaalenii_PYR-1 ILMHADGSARLLEGAHTIALGLRVGWSRPEGRLSIPAGATLMLYTDGLVE
Mflv_4521|M.gilvum_PYR-GCK LVVHADGEVETFDDAHTIALGLRLNWVRPETRRTLEVGSTLVLYTDGLVE
MAV_2108|M.avium_104 IVVHGDGSTQILDEGHTIALGIRRDWPRPEARVTIPARATLLLYTDGLVE
MMAR_3479|M.marinum_M ITTPTEG-------------GTRISWTVPFGDL-----------------
MSMEG_3941|M.smegmatis_MC2_155 ITSSAGS-------------GTHVRWSAPLSGA-----------------
Mb2052c|M.bovis_AF2122/97 VENMPTG-------------GTLLRWSAPLR-------------------
Rv2027c|M.tuberculosis_H37Rv VENMPTG-------------GTLLRWSAPLR-------------------
MAB_3890c|M.abscessus_ATCC_199 VSAMDTG-------------GTRLVWSAPLP-------------------
TH_2781|M.thermoresistible__bu LLAGPGPHATVLTDGASVPLGVQQNRPRPQVSRQLTPGSTLMLYTDGLVE
MUL_3853|M.ulcerans_Agy99 MAEALVS-----------ETHVSHDGAGTTATVVHRLSRPAQFVTDPVVS
: .
Mvan_3797|M.vanbaalenii_PYR-1 RRRTLLEDGISRAAELLEGHRTSDLEDLADQIMSGLAPSVGYQDDVALLL
Mflv_4521|M.gilvum_PYR-GCK RRRIPLEEGISRATDVVVEHRAAPLDDLANAIMSELAPSVGYQDDVALLL
MAV_2108|M.avium_104 RRRSPLDRGIYRVATVAQDARGSSLDELATRIMSAVAPSGGYQDDVVLLL
MMAR_3479|M.marinum_M --------------------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 --------------------------------------------------
Mb2052c|M.bovis_AF2122/97 --------------------------------------------------
Rv2027c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2781|M.thermoresistible__bu RRDSPIDTGMDRACQVLTAMLDAEPEAVADAMLDTLAPADGYDDDVAIVV
MUL_3853|M.ulcerans_Agy99 RAPNRPAIDSEFASTVTESGHIVVRGDVDSNTAPTLDRQIAVES------
Mvan_3797|M.vanbaalenii_PYR-1 YRH-PAPLQLNFPADANHLAPARAALRSWLTSARVEPARVMDVLVAAGEA
Mflv_4521|M.gilvum_PYR-GCK YRH-PAPLELTFPAHADHLAPTRTALRRWLSGAKVEQGRVMEVLVAAGEA
MAV_2108|M.avium_104 YRH-PAPLELNFPADVNQLAPARTALRNWLRRVRLDPEQTLNVLVAAGEA
MMAR_3479|M.marinum_M --------------------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 --------------------------------------------------
Mb2052c|M.bovis_AF2122/97 --------------------------------------------------
Rv2027c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2781|M.thermoresistible__bu YRHTFAPLRIELPATPDRLSEVRARLTGWLTAAGAPELLISDIVLAVNEA
MUL_3853|M.ulcerans_Agy99 -RSGIASLTIDLSAVTHLGSAG------------------VSALVTARDR
Mvan_3797|M.vanbaalenii_PYR-1 LANAIEHGHRHTPDGTVSLRATALVDRVRLTVADTGSWKVPQPGRDSRRG
Mflv_4521|M.gilvum_PYR-GCK VANAIEHGHRHGPPGTVRLRATALVDLVELIITDSGSWKPPSAIPDTYRG
MAV_2108|M.avium_104 VANAIEHGHRHSPQGTIRLGATALGDQVRLTITDTGTWKVPQATSYPHRG
MMAR_3479|M.marinum_M --------------------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 --------------------------------------------------
Mb2052c|M.bovis_AF2122/97 --------------------------------------------------
Rv2027c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2781|M.thermoresistible__bu CTNSVEHAYRDREPGPMQVYADLRDGQIMLQVRDFGTWKTPEPNP-RIRG
MUL_3853|M.ulcerans_Agy99 ARKQGGRCVLMAPPGSPAHHVLSLVQIPVISGETENVFAED---------
Mvan_3797|M.vanbaalenii_PYR-1 RGIALMRALMHDVEIDIGRSGTSVHLDTRID------
Mflv_4521|M.gilvum_PYR-GCK RGIGIMRGLMQDVTIAHDSDGTTIHLYARIS------
MAV_2108|M.avium_104 RGIPLMKSLMNDVDIRSDTGGTTVQLSARIT------
MMAR_3479|M.marinum_M -------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 -------------------------------------
Mb2052c|M.bovis_AF2122/97 -------------------------------------
Rv2027c|M.tuberculosis_H37Rv -------------------------------------
MAB_3890c|M.abscessus_ATCC_199 -------------------------------------
TH_2781|M.thermoresistible__bu RGLPMMRAISADVQVDNTRSGTVVELTFTLSGQPALT
MUL_3853|M.ulcerans_Agy99 -------------------------------------