For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSGDVTDAGSATAVPGGVAGLTPQWLTAVLRTDAGLPDTATVSEVRAERIAADSGFSSLLYRLHLTGGD GLPATLIAKLPAESQARGAMELLGGYRREIAFYRDVAGRAPLDTPRVYAARMAEDSVDFVLLLEDLRDWD NADHLAGLSMDRARLAVANLAALHAWSVEPANAVALQSFPSLATSVVRDLLVPAFAPGWQIYRDKSGAAV PRRVARFAERFAEMAPQALSALTEHSMLLHGDIRADNMFFDGDRLKIVDFQFASVGCGAADIGYLVSQGL PSEVRRGHDESLVREYLGALQDRGLVSYQFDDAWRHYRFAVAYLMVLPVITLIGWDTLPERSRALCLELT ARAVATADDIDAMEVFA
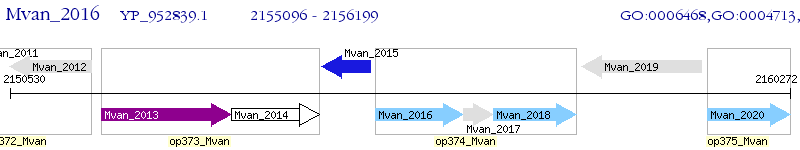
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2016 | - | - | 100% (367) | aminoglycoside phosphotransferase |
| M. vanbaalenii PYR-1 | Mvan_3433 | - | 3e-27 | 26.46% (359) | aminoglycoside phosphotransferase |
| M. vanbaalenii PYR-1 | Mvan_4223 | - | 4e-20 | 28.02% (364) | hypothetical protein Mvan_4223 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4330 | - | 1e-161 | 79.04% (353) | hypothetical protein Mflv_4330 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4035 | - | 1e-36 | 31.88% (367) | hypothetical protein MMAR_4035 |
| M. avium 104 | MAV_1568 | - | 6e-34 | 31.51% (365) | phosphotransferase enzyme family protein, putative |
| M. smegmatis MC2 155 | MSMEG_3876 | - | 9e-32 | 29.57% (345) | phosphotransferase enzyme family protein, putative |
| M. thermoresistible (build 8) | TH_3623 | - | 1e-36 | 30.90% (343) | PUTATIVE putative aminoglycoside phosphotransferase |
| M. ulcerans Agy99 | MUL_1045 | - | 2e-15 | 27.66% (329) | hypothetical protein MUL_1045 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2016|M.vanbaalenii_PYR-1 ---MTSGDVTDAGSATAVPGGVAGLTPQWLTAVLRTDAGLPDTATVSEVR
Mflv_4330|M.gilvum_PYR-GCK ---------------MTVPGSVDALTACWLTEALRSDPTLSDTLTVDGVR
MMAR_4035|M.marinum_M --------------MVSIPRHPTDVTPQWLAAVLGERG--TPVGVSGVDV
MAV_1568|M.avium_104 --------------MLSIPRYPGDVTPQWLSAALSGRG--APVEVAEVDV
MSMEG_3876|M.smegmatis_MC2_155 -----------MTAAVSIPGRPVDVTPSWLSGVLG-------TEVATAAT
TH_3623|M.thermoresistible__bu --------------MAFTDYTIAELTPEALAAVLRRRPGWEALTVTTVES
MUL_1045|M.ulcerans_Agy99 MIALARLATHLGRGLGRVATDAVLRNARAFPRRIHDLDPPALSEIMGRRV
.. :. :
Mvan_2016|M.vanbaalenii_PYR-1 AERIAADSGFSSLLYRLHLTGGDGLPATLIAKLPAESQARGAMELLGG-Y
Mflv_4330|M.gilvum_PYR-GCK AERIAMDSGFSSLLYRLHLTG-TGVPATVIAKLPAESEARGAMELLGG-Y
MMAR_4035|M.marinum_M VPIGTGQTGATYRLTVIYDPDPAGLPDTFVIKLPAQDDTVRDRVVVG--Y
MAV_1568|M.avium_104 AAIGTGQTGATYRVTARYATDPGDLPQTFVIKLPAQDDTVRDRVVIG--Y
MSMEG_3876|M.smegmatis_MC2_155 EPIGTGQTGATYRVTVTYAGDS-ALPTTFAIKLPSQDDTVRDRVAIG--Y
TH_3623|M.thermoresistible__bu QPVGTGQMASSYRLTLGYAGDSGSAPDTLIAKISSTDETSWQMAVATGAY
MUL_1045|M.ulcerans_Agy99 SSVSVLDDSSGTTSRARLALSGNDVPESVFVKMAAATAATRFIGELGRLA
. : . . * :. *:.: :
Mvan_2016|M.vanbaalenii_PYR-1 RREIAFYRDVAGRAPLDTPRVYAARMAEDSVDFVLLLEDLRDWDN-----
Mflv_4330|M.gilvum_PYR-GCK RRELAFYRNVAGRAPLDTPHVYTAQIAEDSVDFVLLLEDLRDWDN-----
MMAR_4035|M.marinum_M RSECAFYSSVADRVRVPTPRCFYCEITEDAMDFALLLEDQAPAVQ-----
MAV_1568|M.avium_104 RSECAFYSSVADRVQVPTPRCFYSEITEDAMDFALLLADQAPAVQ-----
MSMEG_3876|M.smegmatis_MC2_155 RSEHAFYTSLAGQVRIPVPHCYHCEIAGEGADFVLVLADMAPAEQ-----
TH_3623|M.thermoresistible__bu EREVRYYQHLSGLAATRTPECYFAEIADDRCGFALLLEDMGPADT-----
MUL_1045|M.ulcerans_Agy99 ETEVSFYRDLSPELAPDAPTAYGCAFDPLTGRFVLVLEDLAAPAEGTCDF
. * :* :: .* : . : *.*:* *
Mvan_2016|M.vanbaalenii_PYR-1 ADHLAGLSMDRARLAVANLAALHAWSVEPANAVALQSFP-SLATSVVRDL
Mflv_4330|M.gilvum_PYR-GCK ADHLVGLTVEQTRLAVTNLAGLHAWSVDPSNSAALQHFP-SLSTPIVRDL
MMAR_4035|M.marinum_M GDQLTGCGEREARLAVVALAGLHGPSWCDPVWLDQPGIAFPFPDEPFANG
MAV_1568|M.avium_104 GDQLLGCGETEARLAVTALAGLHAPSWCDPVWLDFPGIAFGRPDEAGAGG
MSMEG_3876|M.smegmatis_MC2_155 GDQIAGCSPAEAALAVRALADLHGPTWCDPQWANFPGIAMPKPEPDSAKG
TH_3623|M.thermoresistible__bu IDQISGCTADQADLALGQAAALHGSSWAHAALRDND----WLPAQGAWAK
MUL_1045|M.ulcerans_Agy99 SDTLHPFDNDQAALVVELLARLHATYWVRLNHGGVRPFG--WRYAASRDP
* : .: *.: * **.
Mvan_2016|M.vanbaalenii_PYR-1 LVPAFAPGWQIYRDKSGAAVPRRVARFAERFAEMAPQALSALTEHSMLLH
Mflv_4330|M.gilvum_PYR-GCK LVPAFAPGWQVYRDRSGAAVPRRIARFADRFAELAPQALAALVEHSMLLH
MMAR_4035|M.marinum_M LGEVARMSAEITLDKLGDRLSAADRETFTETMNLVTPWLLAERDRFALLH
MAV_1568|M.avium_104 MGEVAKMSADITLERLGDRMSAEDRETFAAAMGVIAPWLLSEYDRFALLH
MSMEG_3876|M.smegmatis_MC2_155 FGDIAKMAADITLDKLGDRLSIGDRETLSASMSVVTPWLLAEPDRFAVLH
TH_3623|M.thermoresistible__bu LADAIPHITPMWLDRFGAHLRPEHIPVVEQLGDHVSAWLETLTEHRSLWH
MUL_1045|M.ulcerans_Agy99 SVPLVPTMLRASWRRLGGHAAALLPGRDFVFEHYRAITGLIDAPPHTVLH
: * . : *
Mvan_2016|M.vanbaalenii_PYR-1 GDIRADNMFFDGDR----LKIVDFQFASVGCGAADIGYLVSQGLPSEVRR
Mflv_4330|M.gilvum_PYR-GCK GDIRADNMFFDGGR----LKIVDFQFASVGSGAADIAYLVSQGLPTDARR
MMAR_4035|M.marinum_M GDYRLDNLLFDPDL--RRVAVVDWQTLGIGLPARDLGYFTATSLNSQLRS
MAV_1568|M.avium_104 GDYRLDNLLFDPDR--TRVSVVDWQTLGVGLPARDLAYFTATSLDSPLRA
MSMEG_3876|M.smegmatis_MC2_155 GDYRLDNMLFDPDR--TRVTVVDWQTLGSGLPARDLSYFTATSLDPADRA
TH_3623|M.thermoresistible__bu GDFRLDNLLFGAQAGATPIAVVDWQSVASGPGVIDVSYFLGNSMTEDHRA
MUL_1045|M.ulcerans_Agy99 GDAHPGNWYFRSSGGPLAAGLLDWQAIRLGHPARDLAYTLITGMTTTDRR
** : .* * ::*:* * . *:.* .: *
Mvan_2016|M.vanbaalenii_PYR-1 GHDESLVREYLGALQDRGLVSYQFDDAWRHYRFAVAYLMVLPVITLIGWD
Mflv_4330|M.gilvum_PYR-GCK GHDETLMREYLGMLG----VPYRFDDAWRHYRFAVAYLMVLPVIILVGWD
MMAR_4035|M.marinum_M DIEESLVDDYHRKLLSYGVTGYDWETCWRDYRLGMPHTVLISALGFA-FA
MAV_1568|M.avium_104 AIEEELVADYHRALVAGGVNGYDRETCWRDYRLGVLQAPLISALGFA-FA
MSMEG_3876|M.smegmatis_MC2_155 AAERDLVGEYHRTLLAHGVTGYDRETCWRDYRLGMLQAPLITVLGCA-FA
TH_3623|M.thermoresistible__bu EHERELVGEYHRRLLSHGVQGYSADRCWLEYRAHAVFGLMLTVPVSL-GV
MUL_1045|M.ulcerans_Agy99 VTERELLDVYRRALAAAGGPELDREEPWLRYRQAAACAYVATTIAAG---
:. *: * * : * ** : ..
Mvan_2016|M.vanbaalenii_PYR-1 TLPERSRALCLELTARAVATADDIDAMEVFA-------------------
Mflv_4330|M.gilvum_PYR-GCK SLPERSRALCLELTSRAIAAIDDVDATEVFA-------------------
MMAR_4035|M.marinum_M TATERGDEMVLTMLGRGCQAIRDLETLELIG-------------------
MAV_1568|M.avium_104 AATERGDDMVLTMLGRGCRAIRELGTLELIG-------------------
MSMEG_3876|M.smegmatis_MC2_155 SSTDRGDDMMVVMAQRGCQAIRELGTLDLIDAAS----------------
TH_3623|M.thermoresistible__bu QATERGDKMFAAMATRAAAQILENESYTALKCL-----------------
MUL_1045|M.ulcerans_Agy99 LGGMQAADIALTGLDLTVAALHDLQTVAALRAADLGGTSWAWRPGMARSA
:. : : : :
Mvan_2016|M.vanbaalenii_PYR-1 ---------------------------------
Mflv_4330|M.gilvum_PYR-GCK ---------------------------------
MMAR_4035|M.marinum_M ---------------------------------
MAV_1568|M.avium_104 ---------------------------------
MSMEG_3876|M.smegmatis_MC2_155 ---------------------------------
TH_3623|M.thermoresistible__bu ---------------------------------
MUL_1045|M.ulcerans_Agy99 RIRPGASARRCPMVWSTSSHILGVSRHFAMGAW