For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAAVSIPGRPVDVTPSWLSGVLGTEVATAATEPIGTGQTGATYRVTVTYAGDSALPTTFAIKLPSQDDT VRDRVAIGYRSEHAFYTSLAGQVRIPVPHCYHCEIAGEGADFVLVLADMAPAEQGDQIAGCSPAEAALAV RALADLHGPTWCDPQWANFPGIAMPKPEPDSAKGFGDIAKMAADITLDKLGDRLSIGDRETLSASMSVVT PWLLAEPDRFAVLHGDYRLDNMLFDPDRTRVTVVDWQTLGSGLPARDLSYFTATSLDPADRAAAERDLVG EYHRTLLAHGVTGYDRETCWRDYRLGMLQAPLITVLGCAFASSTDRGDDMMVVMAQRGCQAIRELGTLDL IDAAS
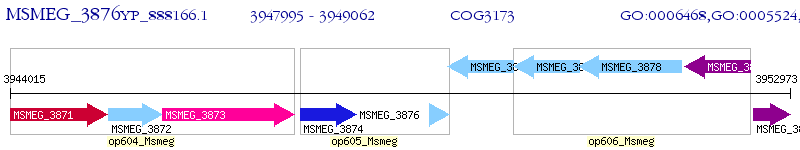
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3876 | - | - | 100% (355) | phosphotransferase enzyme family protein, putative |
| M. smegmatis MC2 155 | MSMEG_3846 | - | 2e-36 | 31.62% (351) | phosphotransferase enzyme family protein |
| M. smegmatis MC2 155 | MSMEG_4885 | - | 1e-25 | 29.23% (366) | hypothetical protein MSMEG_4885 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3102 | - | 1e-132 | 64.29% (350) | aminoglycoside phosphotransferase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4589c | - | 2e-07 | 40.85% (71) | putative phosphotransferase |
| M. marinum M | MMAR_4035 | - | 1e-131 | 64.59% (353) | hypothetical protein MMAR_4035 |
| M. avium 104 | MAV_1568 | - | 1e-135 | 66.01% (353) | phosphotransferase enzyme family protein, putative |
| M. thermoresistible (build 8) | TH_0441 | - | 1e-142 | 67.04% (361) | phosphotransferase enzyme family protein, putative |
| M. ulcerans Agy99 | MUL_1045 | - | 2e-18 | 25.82% (364) | hypothetical protein MUL_1045 |
| M. vanbaalenii PYR-1 | Mvan_3433 | - | 1e-137 | 67.24% (348) | aminoglycoside phosphotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3102|M.gilvum_PYR-GCK ---------------------------MTIPRTPADLTPTWLESVLDAG-
Mvan_3433|M.vanbaalenii_PYR-1 ---------------------------MTIPRSPQDITPTWLASVLGTE-
MSMEG_3876|M.smegmatis_MC2_155 -----------------------MTAAVSIPGRPVDVTPSWLSGVLGTE-
TH_0441|M.thermoresistible__bu -----------------------MSTVLSIPSGPAEVTPEWLSTVLSADG
MMAR_4035|M.marinum_M --------------------------MVSIPRHPTDVTPQWLAAVLGERG
MAV_1568|M.avium_104 --------------------------MLSIPRYPGDVTPQWLSAALSGRG
MUL_1045|M.ulcerans_Agy99 MIALARLATHLGRGLGRVATDAVLRNARAFPRRIHDLDPPALSEIMGRR-
MAB_4589c|M.abscessus_ATCC_199 --------------MHKDVSSDSDGPTGDAASLDADAIPTSLSEYLNREL
. : * * :.
Mflv_3102|M.gilvum_PYR-GCK ----VDDVTVTAIGTGQTGATYRVSATYDNPTAADLPATFAVKLPAQDDA
Mvan_3433|M.vanbaalenii_PYR-1 ----VSGVEVAAIGTGQTGATYRVTATY--PESPSPPATFAVKLPAQDDA
MSMEG_3876|M.smegmatis_MC2_155 ----VATAATEPIGTGQTGATYRVTVTY-AGD-SALPTTFAIKLPSQDDT
TH_0441|M.thermoresistible__bu TPVRVRSVDVVPIGTGQTGATYRLTVGY-ETERADLPPTFAIKLPAQDES
MMAR_4035|M.marinum_M TPVGVSGVDVVPIGTGQTGATYRLTVIY-DPDPAGLPDTFVIKLPAQDDT
MAV_1568|M.avium_104 APVEVAEVDVAAIGTGQTGATYRVTARY-ATDPGDLPQTFVIKLPAQDDT
MUL_1045|M.ulcerans_Agy99 ----VSSVSVLDDSSGTTSRARLALSGN------DVPESVFVKMAAATAA
MAB_4589c|M.abscessus_ATCC_199 G----EQVSVRRLSAGHSNLTYVVTGSS-GTQWILRRPPFGRLQAGAHDV
. . .:* :. : .. ..
Mflv_3102|M.gilvum_PYR-GCK VRERVALG--YLSEVDFYSTITADVAVPVPGCYHSEISSDGTDFVLVLAD
Mvan_3433|M.vanbaalenii_PYR-1 VRERVALG--YLSEVDFYAAINRDVAIPVPDCFHTEISTDGKDFVLVLAD
MSMEG_3876|M.smegmatis_MC2_155 VRDRVAIG--YRSEHAFYTSLAGQVRIPVPHCYHCEIAGEGADFVLVLAD
TH_0441|M.thermoresistible__bu VRERVALG--YRAEHAFYTRVAAHMAIPVPHCYHCEIGGDGAEFAMLMAD
MMAR_4035|M.marinum_M VRDRVVVG--YRSECAFYSSVADRVRVPTPRCFYCEITEDAMDFALLLED
MAV_1568|M.avium_104 VRDRVVIG--YRSECAFYSSVADRVQVPTPRCFYSEITEDAMDFALLLAD
MUL_1045|M.ulcerans_Agy99 TRFIGELGRLAETEVSFYRDLSPELAPDAPTAYGCAFDPLTGRFVLVLED
MAB_4589c|M.abscessus_ATCC_199 MREYRVLD----------ALSTAQVRVPRVTLASTDAEIFGAPFYLMERQ
* :. : * :: :
Mflv_3102|M.gilvum_PYR-GCK MAPAEQG-----DQIAGCTAAEAALAVEAIAGLHGPSWGDRRYFDLPSIV
Mvan_3433|M.vanbaalenii_PYR-1 MAPAEQG-----DQIAGCTPAEAALAVEALAGLHGPSWNERRYFDLPSIV
MSMEG_3876|M.smegmatis_MC2_155 MAPAEQG-----DQIAGCSPAEAALAVRALADLHGPTWCDPQWANFPGIA
TH_0441|M.thermoresistible__bu MAPAVQG-----DQIVGCSPAEARLAVTALAGLHGPSWCDPQWADVPGIA
MMAR_4035|M.marinum_M QAPAVQG-----DQLTGCGEREARLAVVALAGLHGPSWCDPVWLDQPGIA
MAV_1568|M.avium_104 QAPAVQG-----DQLLGCGETEARLAVTALAGLHAPSWCDPVWLDFPGIA
MUL_1045|M.ulcerans_Agy99 LAAPAEGTCDFSDTLHPFDNDQAALVVELLARLHATYWVRLNHGGVRPFG
MAB_4589c|M.abscessus_ATCC_199 DGEVIRD----VSPEWFVGSARRELVLDLAVALAEIHNADP-----TRLV
. .. . . *.: . * :
Mflv_3102|M.gilvum_PYR-GCK MPKPGDEAAASGMGEVAVMAANMTLEKLGGSVDDEDRDTLLTSMSLVTPW
Mvan_3433|M.vanbaalenii_PYR-1 MPKPGDEAAAAGMGEVAKMAADITLDKLGGSVSDEDRDTLVTSMSLVTPW
MSMEG_3876|M.smegmatis_MC2_155 MPKP-EPDSAKGFGDIAKMAADITLDKLGDRLSIGDRETLSASMSVVTPW
TH_0441|M.thermoresistible__bu MPKP-DDAAAEGLGEIAEMAADITIDKLGPRMRPEDRETTLTAMSMVTAW
MMAR_4035|M.marinum_M FPFP-DEPFANGLGEVARMSAEITLDKLGDRLSAADRETFTETMNLVTPW
MAV_1568|M.avium_104 FGRP-DEAGAGGMGEVAKMSADITLERLGDRMSAEDRETFAAAMGVIAPW
MUL_1045|M.ulcerans_Agy99 WRYAASRDPSVPLVPTMLRASWRRLGGHAAALLPG-RDFVFEHYRAITGL
MAB_4589c|M.abscessus_ATCC_199 EAKLGRESGYLARQLKTWGGQWESIKELPTGRDLQDHTTITAWLTEN---
. : :
Mflv_3102|M.gilvum_PYR-GCK LMAVPDRFSLMHGDYRLDNLLYDPE--RTRVTVVDWQTVGIGLPTRDLAY
Mvan_3433|M.vanbaalenii_PYR-1 LLAVPDRFSLMHGDYRLDNLLYDPD--RTRVTVVDWQTMGIGLPSRDLAY
MSMEG_3876|M.smegmatis_MC2_155 LLAEPDRFAVLHGDYRLDNMLFDPD--RTRVTVVDWQTLGSGLPARDLSY
TH_0441|M.thermoresistible__bu LRTEKSRFALMHGDYRLDNMLFDPD--RTRVTVVDWQTMGAGLPARDLSY
MMAR_4035|M.marinum_M LLAERDRFALLHGDYRLDNLLFDPD--LRRVAVVDWQTLGIGLPARDLGY
MAV_1568|M.avium_104 LLSEYDRFALLHGDYRLDNLLFDPD--RTRVSVVDWQTLGVGLPARDLAY
MUL_1045|M.ulcerans_Agy99 IDAPP--HTVLHGDAHPGNWYFRSSGGPLAAGLLDWQAIRLGHPARDLAY
MAB_4589c|M.abscessus_ATCC_199 -YPGDRPAAVVHGDYKLDNVLVDPAT-ARITAVLDWEMATVGDPLADLGY
. :::*** : .* . . ::**: * * **.*
Mflv_3102|M.gilvum_PYR-GCK FTATSLLPD--------DRAAVERDLVQRYHAALLAHGVSGYDVETCWQD
Mvan_3433|M.vanbaalenii_PYR-1 FTATSLLPE--------TRAEVERDLVEHYHAALLGHGVSGYDVETCWHD
MSMEG_3876|M.smegmatis_MC2_155 FTATSLDPA--------DRAAAERDLVGEYHRTLLAHGVTGYDRETCWRD
TH_0441|M.thermoresistible__bu FLGTSLLPE--------VRSEVERDLVEVYHRALRSHGVADYDSETCWRD
MMAR_4035|M.marinum_M FTATSLNSQ--------LRSDIEESLVDDYHRKLLSYGVTGYDWETCWRD
MAV_1568|M.avium_104 FTATSLDSP--------LRAAIEEELVADYHRALVAGGVNGYDRETCWRD
MUL_1045|M.ulcerans_Agy99 TLITGMTTT--------DRRVTERELLDVYRRALAAAGGPELDREEPWLR
MAB_4589c|M.abscessus_ATCC_199 LLYMTPEPGGEVAMSELTGSVTAQEGCPSHTEIAQAYREARSGDQSYWTP
. .. : . . : *
Mflv_3102|M.gilvum_PYR-GCK YRLGVFQAP----LITALGFAFAASTERGDE-MILTMLHRGCRAIRELGA
Mvan_3433|M.vanbaalenii_PYR-1 YRLGVLQAP----LITVLGTAFASSTERGDE-MMLTMLHRGCRAIRELAS
MSMEG_3876|M.smegmatis_MC2_155 YRLGMLQAP----LITVLGCAFASSTDRGDD-MMVVMAQRGCQAIRELGT
TH_0441|M.thermoresistible__bu YRLGMLQVP----LLTVLGFAFAASTERGDD-MMLTMLERGGRAIRELDS
MMAR_4035|M.marinum_M YRLGMPHTV----LISALGFAFATATERGDE-MVLTMLGRGCQAIRDLET
MAV_1568|M.avium_104 YRLGVLQAP----LISALGFAFAAATERGDD-MVLTMLGRGCRAIRELGT
MUL_1045|M.ulcerans_Agy99 YRQAAACAY----VATTIAAGLGG--MQAAD-IALTGLDLTVAALHDLQT
MAB_4589c|M.abscessus_ATCC_199 EDLVYYQVLGGWKLATLLEVSYQRHLAGTTDDPFFAELDEGVPRLLSVAR
. : : : . : .. : .:
Mflv_3102|M.gilvum_PYR-GCK IDLVRSYQASTQPSSPTTA-------------------------------
Mvan_3433|M.vanbaalenii_PYR-1 IDLVRSYQVS----SPTTA-------------------------------
MSMEG_3876|M.smegmatis_MC2_155 LDLIDAAS------------------------------------------
TH_0441|M.thermoresistible__bu LELVNPAAS-----------------------------------------
MMAR_4035|M.marinum_M LELIG---------------------------------------------
MAV_1568|M.avium_104 LELIG---------------------------------------------
MUL_1045|M.ulcerans_Agy99 VAALRAADLGGTSWAWRPGMARSARIRPGASARRCPMVWSTSSHILGVSR
MAB_4589c|M.abscessus_ATCC_199 SLIPAAG-------------------------------------------
Mflv_3102|M.gilvum_PYR-GCK -------
Mvan_3433|M.vanbaalenii_PYR-1 -------
MSMEG_3876|M.smegmatis_MC2_155 -------
TH_0441|M.thermoresistible__bu -------
MMAR_4035|M.marinum_M -------
MAV_1568|M.avium_104 -------
MUL_1045|M.ulcerans_Agy99 HFAMGAW
MAB_4589c|M.abscessus_ATCC_199 -------