For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSETALLVIDMFNTYTHPDAEQLADNVVEVVDPIAELVARAAERDDVDLVYVNDNYGDFTATHSDVVRAA LEGARPELVRPLVPASGSRFLTKVRHSAFYATPLDYLLSRLGVRRIVLTGQVTEQCILYTALDGYVRHYD VVVPPDAVAHIDSQLGEAALEMMRRNMKAELSKSTECLP
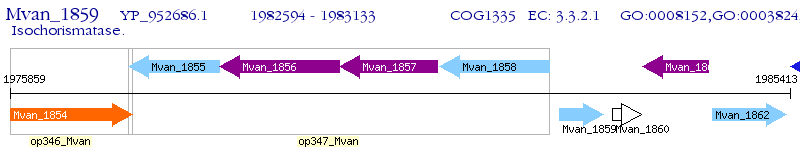
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1859 | - | - | 100% (179) | isochorismatase hydrolase |
| M. vanbaalenii PYR-1 | Mvan_3053 | - | 2e-10 | 27.81% (187) | isochorismatase hydrolase |
| M. vanbaalenii PYR-1 | Mvan_4942 | - | 1e-06 | 31.03% (145) | isochorismatase hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4511 | - | 5e-78 | 78.21% (179) | isochorismatase hydrolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2557c | - | 2e-67 | 67.98% (178) | putative isochorismatase hydrolase EntB |
| M. marinum M | MMAR_3790 | - | 1e-05 | 38.98% (59) | isochorismatase family protein |
| M. avium 104 | MAV_1736 | - | 3e-63 | 64.94% (174) | isochorismatase family protein |
| M. smegmatis MC2 155 | MSMEG_4618 | - | 2e-68 | 64.61% (178) | isochorismatase family protein |
| M. thermoresistible (build 8) | TH_2198 | - | 3e-72 | 70.39% (179) | isochorismatase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1859|M.vanbaalenii_PYR-1 ------------------------------------------MSETALLV
Mflv_4511|M.gilvum_PYR-GCK ------------------------------------MDISSAMSETALLV
MSMEG_4618|M.smegmatis_MC2_155 ------------------------------------------MSDTAVVI
TH_2198|M.thermoresistible__bu ------------------------------------------MSDTAVVV
MAV_1736|M.avium_104 -----------------------------------------------MLV
MAB_2557c|M.abscessus_ATCC_199 ----------------------------MDLVATAAQRVPQGMSETALLV
MMAR_3790|M.marinum_M MPEYFSDTSRWTCSPEHVDLDIPRRFTRRVRLAAQPQHVTLDLGDAAVIV
:::
Mvan_1859|M.vanbaalenii_PYR-1 IDMFNTYTHPDAEQLADNVVE-----VVDPIAELVARAAERDDVDLVYVN
Mflv_4511|M.gilvum_PYR-GCK IDMFNTYDHPDAEPLADNVAA-----IVDPLTELIRRSNERDDVAVVYVN
MSMEG_4618|M.smegmatis_MC2_155 VDMLNTYEHEDAELLAPNVEK-----IIDPLVKLIGEARDREDVDLIYVN
TH_2198|M.thermoresistible__bu VDMFNTYDHEDAELLTPNVES-----ILDPLAKLIAEARHRDDVDLIYVN
MAV_1736|M.avium_104 VDMMNSYQHPDAENLIPNVEK-----IIEPLTGLVRRARESAGVDLVYVN
MAB_2557c|M.abscessus_ATCC_199 IDMFNTYRHPDADQLAANVAT-----IVEPLAGLVSRAQAHTDMDLIYVN
MMAR_3790|M.marinum_M VDMQNDFCHERGWLASQGVDVRGGAGLKTPINAMLDGFRG-AQVPIIWLN
:** * : * . .* : *: :: : ::::*
Mvan_1859|M.vanbaalenii_PYR-1 DNYGDFTATHSDVVRAALEGARPE---------LVRPLVPASG-------
Mflv_4511|M.gilvum_PYR-GCK DNYGDFAAEPSDIVEAAVNGARPE---------LVRPLVPGPE-------
MSMEG_4618|M.smegmatis_MC2_155 DNYGDFTAQFSDIVADALDGKRPD---------LVRPILPYDG-------
TH_2198|M.thermoresistible__bu DNHGDFTAEFSDVVRTALDGARPD---------LVRPIVPDDG-------
MAV_1736|M.avium_104 DNYGDFTAQFSDLVRSALDGARPD---------LVKPIAPVSGE------
MAB_2557c|M.abscessus_ATCC_199 DNHGDFAAGPSAIVGAALHGAHPE---------LVRPLVPEGE-------
MMAR_3790|M.marinum_M WGTRSDRANLPPNVLHAYDGEGTGGGIGESVSLLSNAVLTANSWGAAILD
. . * . * * .* . * ..: .
Mvan_1859|M.vanbaalenii_PYR-1 -------SRFLTKVRHSAFYATPLDYLLSRLGVRRIVLTGQVTEQCILYT
Mflv_4511|M.gilvum_PYR-GCK -------ARFLTKVRHSAFYATSLDYLLTRLDTRRIILTGQVTEQCVLYT
MSMEG_4618|M.smegmatis_MC2_155 -------CRLLTKVRHSVFYATALDYLLGRLGAKRVILAGQVTEQCILYS
TH_2198|M.thermoresistible__bu -------CRALLKVRHSVFYATALEYLLTRLSARRVILTGQVTEQCILYS
MAV_1736|M.avium_104 -------AASLTKVRHSAFYSTALAYLLSRLGTKRLIITGQVTEQCILYT
MAB_2557c|M.abscessus_ATCC_199 -------FQFVTKVRHSIFYASPLEYLLGRLRTRQLILCGQVTEQCVLYS
MMAR_3790|M.marinum_M GLVVDDIDIHVNKHRMSGFWDTELDSILRNLGVRHLFLCGVNVDQCVYAT
: * * * *: : * :* .* .:::.: * .:**: :
Mvan_1859|M.vanbaalenii_PYR-1 ALDGYVRHYDVVVPPDAVAHIDSQLGEAALEMMRRN----------MKAE
Mflv_4511|M.gilvum_PYR-GCK ALDGYVRHYDVVVPPDTVAHIDDKLGTAALEMMERN----------MKAE
MSMEG_4618|M.smegmatis_MC2_155 ALDAYVRHFELVVPSDAVAHIDAELGDAALEMMRRN----------MSAD
TH_2198|M.thermoresistible__bu ALDAYVRHFEVVVPPDAVAHIDAGLGEAALQMMARN----------MSAE
MAV_1736|M.avium_104 ALDAYVRHFPVVIPTDAVAHIDPELGAAACKMMEQN----------MSAE
MAB_2557c|M.abscessus_ATCC_199 ALDAYVRHFDVVIPTDAVAPIDPSLGESAVEMMRRN----------MRAE
MMAR_3790|M.marinum_M LIDAACAGYDCLLITDASATTSPSYCTEATIYNVRQCFGFTCDTAAIRAS
:*. : :: .*: * . * :: : *.
Mvan_1859|M.vanbaalenii_PYR-1 LSKSTECLP--------
Mflv_4511|M.gilvum_PYR-GCK LPESTASLP--------
MSMEG_4618|M.smegmatis_MC2_155 VIPAVDCLS--------
TH_2198|M.thermoresistible__bu LPPSADCLP--------
MAV_1736|M.avium_104 LTTAADCLG--------
MAB_2557c|M.abscessus_ATCC_199 LCTSTRCIH--------
MMAR_3790|M.marinum_M LHAPTEPHGVCGTCQPE
: ..