For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DISSAMSETALLVIDMFNTYDHPDAEPLADNVAAIVDPLTELIRRSNERDDVAVVYVNDNYGDFAAEPSD IVEAAVNGARPELVRPLVPGPEARFLTKVRHSAFYATSLDYLLTRLDTRRIILTGQVTEQCVLYTALDGY VRHYDVVVPPDTVAHIDDKLGTAALEMMERNMKAELPESTASLP*
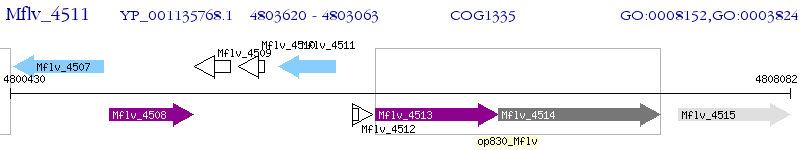
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4511 | - | - | 100% (185) | isochorismatase hydrolase |
| M. gilvum PYR-GCK | Mflv_2275 | - | 5e-06 | 27.27% (132) | isochorismatase hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2557c | - | 1e-63 | 66.48% (179) | putative isochorismatase hydrolase EntB |
| M. marinum M | MMAR_2657 | - | 2e-05 | 29.89% (87) | isochorismatase family protein |
| M. avium 104 | MAV_1736 | - | 6e-59 | 64.07% (167) | isochorismatase family protein |
| M. smegmatis MC2 155 | MSMEG_4618 | - | 4e-61 | 63.16% (171) | isochorismatase family protein |
| M. thermoresistible (build 8) | TH_2198 | - | 2e-67 | 67.04% (179) | isochorismatase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1859 | - | 5e-78 | 78.21% (179) | isochorismatase hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4511|M.gilvum_PYR-GCK --------MDISSAMSETALLVIDMFNTYDHPDAEPLADNVAAIVDPLTE
Mvan_1859|M.vanbaalenii_PYR-1 --------------MSETALLVIDMFNTYTHPDAEQLADNVVEVVDPIAE
MAB_2557c|M.abscessus_ATCC_199 MDLVATAAQRVPQGMSETALLVIDMFNTYRHPDADQLAANVATIVEPLAG
MSMEG_4618|M.smegmatis_MC2_155 --------------MSDTAVVIVDMLNTYEHEDAELLAPNVEKIIDPLVK
TH_2198|M.thermoresistible__bu --------------MSDTAVVVVDMFNTYDHEDAELLTPNVESILDPLAK
MAV_1736|M.avium_104 -------------------MLVVDMMNSYQHPDAENLIPNVEKIIEPLTG
MMAR_2657|M.marinum_M --------------------MGYDLSLVRAPLGPAQTVLEAARKLG-LLI
: *: .. :. : :
Mflv_4511|M.gilvum_PYR-GCK LIRRSNERDDVAVVYVNDNYG----DFAAEPSDIVEAAVNGARP--ELVR
Mvan_1859|M.vanbaalenii_PYR-1 LVARAAERDDVDLVYVNDNYG----DFTATHSDVVRAALEGARP--ELVR
MAB_2557c|M.abscessus_ATCC_199 LVSRAQAHTDMDLIYVNDNHG----DFAAGPSAIVGAALHGAHP--ELVR
MSMEG_4618|M.smegmatis_MC2_155 LIGEARDREDVDLIYVNDNYG----DFTAQFSDIVADALDGKRP--DLVR
TH_2198|M.thermoresistible__bu LIAEARHRDDVDLIYVNDNHG----DFTAEFSDVVRTALDGARP--DLVR
MAV_1736|M.avium_104 LVRRARESAGVDLVYVNDNYG----DFTAQFSDLVRSALDGARP--DLVK
MMAR_2657|M.marinum_M VHTREGHRPDLSDLPANKRWRSARIGAEIGVAGPCGRVLTRGEPGWEIIP
: . .: : .*.. . : .: .* :::
Mflv_4511|M.gilvum_PYR-GCK PLVPGPE-ARFLTKVRHSAFYATSLDYLLTRLDTRRIILTGQVTEQCVLY
Mvan_1859|M.vanbaalenii_PYR-1 PLVPASG-SRFLTKVRHSAFYATPLDYLLSRLGVRRIVLTGQVTEQCILY
MAB_2557c|M.abscessus_ATCC_199 PLVPEGE-FQFVTKVRHSIFYASPLEYLLGRLRTRQLILCGQVTEQCVLY
MSMEG_4618|M.smegmatis_MC2_155 PILPYDG-CRLLTKVRHSVFYATALDYLLGRLGAKRVILAGQVTEQCILY
TH_2198|M.thermoresistible__bu PIVPDDG-CRALLKVRHSVFYATALEYLLTRLSARRVILTGQVTEQCILY
MAV_1736|M.avium_104 PIAPVSGEAASLTKVRHSAFYSTALAYLLSRLGTKRLIITGQVTEQCILY
MMAR_2657|M.marinum_M EMEPLPG-EMVVDKLGKGSFYATDLELILTTRRITHLIFTGIATDVCVHT
: * : *: :. **:: * :* :::: * .*: *:
Mflv_4511|M.gilvum_PYR-GCK TALDGYVRHYDVVVPPDTVAHIDDKLGTAALEMMERN--------MKAEL
Mvan_1859|M.vanbaalenii_PYR-1 TALDGYVRHYDVVVPPDAVAHIDSQLGEAALEMMRRN--------MKAEL
MAB_2557c|M.abscessus_ATCC_199 SALDAYVRHFDVVIPTDAVAPIDPSLGESAVEMMRRN--------MRAEL
MSMEG_4618|M.smegmatis_MC2_155 SALDAYVRHFELVVPSDAVAHIDAELGDAALEMMRRN--------MSADV
TH_2198|M.thermoresistible__bu SALDAYVRHFEVVVPPDAVAHIDAGLGEAALQMMARN--------MSAEL
MAV_1736|M.avium_104 TALDAYVRHFPVVIPTDAVAHIDPELGAAACKMMEQN--------MSAEL
MMAR_2657|M.marinum_M TMREANDRGYECLLLSDCTGATDYANHLAALKMITMQGGVFGAHATNGAL
: :. * : :: .* .. * :* :*: : . :
Mflv_4511|M.gilvum_PYR-GCK PESTASLP----
Mvan_1859|M.vanbaalenii_PYR-1 SKSTECLP----
MAB_2557c|M.abscessus_ATCC_199 CTSTRCIH----
MSMEG_4618|M.smegmatis_MC2_155 IPAVDCLS----
TH_2198|M.thermoresistible__bu PPSADCLP----
MAV_1736|M.avium_104 TTAADCLG----
MMAR_2657|M.marinum_M LEAFNKVRDRRG
: :