For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKRPIPRTVRFGLLSTYPPTPCRLANYSSALFGALSARGSQVSVVRVADGSQSSDARIVGELVNGSARSA AHCVDSLNHSDVAVIQHDYGVYGGAHGDGLLDVIDGLRVPTVAVAHTILKNPAPHQRWVMERMAATIDRM VVMSEAARERLCREYGVDRRKVVTIPYGAVLPTGPRAKRGSRPTILTCGLLGPGKGVERVIDVMSSLQSV PGHPRYVVAGRTHPKVLARDGEAYREARIEQARRLGVADSVTFEDRHLDRASLAALFQAAAVIVLPYDST DQVTSGALVDAVASGRPVVATAFPHAVEVLRDGAGILVPHDDPEALSCALRRVLTQPRLAGSLAAEARQL APAMAWPVVADTYLELAARLLTERQLRV
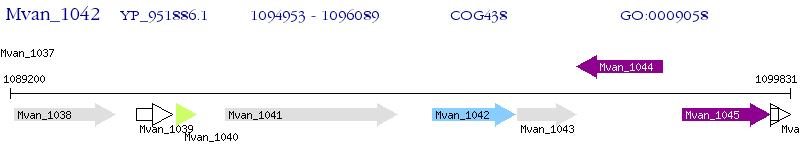
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1042 | - | - | 100% (378) | glycosyl transferase, group 1 |
| M. vanbaalenii PYR-1 | Mvan_1052 | - | e-122 | 61.26% (364) | glycosyl transferase, group 1 |
| M. vanbaalenii PYR-1 | Mvan_5248 | - | 1e-12 | 30.70% (228) | glycosyl transferase, group 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3058 | - | 2e-08 | 26.67% (270) | transferase |
| M. gilvum PYR-GCK | Mflv_5224 | - | 1e-151 | 71.35% (370) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv3032 | - | 2e-08 | 26.67% (270) | transferase |
| M. leprae Br4923 | MLBr_02583 | - | 4e-08 | 26.19% (210) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_1019c | - | 3e-11 | 27.01% (374) | glycosyl transferase |
| M. marinum M | MMAR_0474 | - | 9e-09 | 27.57% (214) | glycosyltransferase |
| M. avium 104 | MAV_4082 | - | 8e-16 | 32.97% (273) | transferase |
| M. smegmatis MC2 155 | MSMEG_1207 | - | 1e-127 | 61.43% (363) | glycosyltransferase, group I |
| M. thermoresistible (build 8) | TH_3188 | - | 3e-13 | 29.17% (264) | PUTATIVE glycosyl transferase, group 1 |
| M. ulcerans Agy99 | MUL_1124 | - | 2e-09 | 28.04% (214) | glycosyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_4082|M.avium_104 ---MRIAMISEHASPLAHLGGVDAGGQNVHVAELSCALARRGHDVVVYTR
TH_3188|M.thermoresistible__bu -------MVSEHASPLAALGGVDAGGQNVHVAELSAALTRRGHEVTVYTR
Mb3058|M.bovis_AF2122/97 --------MRILMVSWEYPPVVIGG-LGRHVHHLSTALAAAGHDVVVLSR
Rv3032|M.tuberculosis_H37Rv --------MRILMVSWEYPPVVIGG-LGRHVHHLSTALAAAGHDVVVLSR
MMAR_0474|M.marinum_M ----MSDLRSVLLLCWRDTGHPQGGGSETYLQRIGAQLAASGIEVTLRTA
MUL_1124|M.ulcerans_Agy99 ----MSDLRSVLLLCWRDTGHPQGGGSETYLQRIGAQLAASGIEVTLRTA
MLBr_02583|M.leprae_Br4923 MFAARRDLGSVLLLCWRDIGHPQGGGSETYLQRIGVQLAASGISVTLRTA
Mvan_1042|M.vanbaalenii_PYR-1 -----MKRPIP---------------RTVRFGLLSTYPPTPCRLANYSSA
Mflv_5224|M.gilvum_PYR-GCK -----MNASVPTFESILAPSARRWTSGRVRFGLLGSYPPSTSGVATFGSA
MSMEG_1207|M.smegmatis_MC2_155 -----MTTASAWPLTP-GVATRQGCSDGLNVGILSTYAPTACGLATFSAA
MAB_1019c|M.abscessus_ATCC_199 -------------MTGSEWFSSLPGGLNRYFTDLFTALRTRGDIELSAAA
. : :
MAV_4082|M.avium_104 RDDP----RLPDSVETPHGYSVVHVPAGPARVLPKDELLQYMYGFAQFLR
TH_3188|M.thermoresistible__bu RDDR----GLPDRVDTPLGYTVVHVPAGPAEQLPKDELLPYMGPFAQFLD
Mb3058|M.bovis_AF2122/97 CPSGTDPSTHPSSDEVTEGVRVIAAAQDPHEFTFGNDMMAWTLAMGHAMI
Rv3032|M.tuberculosis_H37Rv CPSGTDPSTHPSSDEVTEGVRVIAAAQDPHEFTFGNDMMAWTLAMGHAMI
MMAR_0474|M.marinum_M RYPG------APRWELVDGVRVNRAGGR-------YSVYVWALLAMVAAR
MUL_1124|M.ulcerans_Agy99 RYPG------APRWEVVDGVRVNRAGGR-------YSVYVWALLAMVAAR
MLBr_02583|M.leprae_Br4923 RYPG------ASRREVVDGVLISRAGGR-------YSVYLWALLAMAAAR
Mvan_1042|M.vanbaalenii_PYR-1 LFGA-----LSARGSQVSVVRVADGSQSS------DARIVGELVNGSARS
Mflv_5224|M.gilvum_PYR-GCK LAAA-----LTARGSEVSVVRIADGSESP------DAGVIGELVKGSPQS
MSMEG_1207|M.smegmatis_MC2_155 LAGG-----LSGTGANVNVVRIADGAPSG------NARVVGELVNGSAAS
MAB_1019c|M.abscessus_ATCC_199 FGVP------------SDGGSSWGGTGGS----------TRSRVLAAFND
MAV_4082|M.avium_104 QQ---------WCSDGPDVAHAHFWMSGVATQQAARAHSIPTVQTFHALG
TH_3188|M.thermoresistible__bu AE---------WSAHRPDVVHAHFWMSGIATQLASRHLDLPAVQTFHALG
Mb3058|M.bovis_AF2122/97 RAGLRLKKLGTDRSWRPDVVHAHDWLVAHPAIALAQFYDVPMVSTIHATE
Rv3032|M.tuberculosis_H37Rv RAGLRLKKLGTDRSWRPDVVHAHDWLVAHPAIALAQFYDVPMVSTIHATE
MMAR_0474|M.marinum_M MG------LGPLRRVRPAAVIDTQNGLPFLARLVYGRRVVVLVHHCHR-E
MUL_1124|M.ulcerans_Agy99 MG------LGPLQRVRPAAVIDTQNGLPFLARLVYGRRVVVLVHHCHR-E
MLBr_02583|M.leprae_Br4923 FG------LGPLRRVRPDVVVDTQNGWPFLARLLYGSRVVVLVHHCHR-E
Mvan_1042|M.vanbaalenii_PYR-1 AA---------HCVDSLNHSDVAVIQHDYGVYGGAHGDGLLDVIDGLRVP
Mflv_5224|M.gilvum_PYR-GCK VT---------ACVESLNGSEVAVIQHQQGLYGGTHGDEVLAILGGLRVP
MSMEG_1207|M.smegmatis_MC2_155 TA---------ACVELLNQNHVAIVQHEYGIYGGADGDEVVGIIGALDVP
MAB_1019c|M.abscessus_ATCC_199 RS----------GLQQPGILDRHFCLYGPAAIGWGTRSRLVVHFHGPWAA
:
MAV_4082|M.avium_104 LTKRLHQGSDDTSPDCRIDVEARVAR-EADWVAATSTDEVFELVRMGRAR
TH_3188|M.thermoresistible__bu VVKKRHQGAEDTSPQERLQLEAMVAR-GANWVAATCTDEVFELMRMGRSR
Mb3058|M.bovis_AF2122/97 AGRHSGWVSGALSRQVHAVESWLVRE-SDSLITCSASMNDEITELFGPGL
Rv3032|M.tuberculosis_H37Rv AGRHSGWVSGALSRQVHAVESWLVRE-SDSLITCSASMNDEITELFGPGL
MMAR_0474|M.marinum_M QWPVAGPVLGRLGWFVESKLSPWLNR-RNQYVTVSLPSARDLVALG-VDN
MUL_1124|M.ulcerans_Agy99 QWPVAGPVLGRLGWFVESKLSPWLNR-RNQYVTVSLPSARDLVALG-VDN
MLBr_02583|M.leprae_Br4923 QWPVAGPVIGRLGWYVESTLSPRLHR-RNQYLTVSLPSARDLVALG-VDH
Mvan_1042|M.vanbaalenii_PYR-1 TVAVAHTILKNPAPHQRWVMERMAAT-IDRMVVMSEAARERLCREYGVDR
Mflv_5224|M.gilvum_PYR-GCK SIVIVHDVSKSPEPHQRWVLEQIVTM-ADSVVVMSEAARARLRGEYGVDG
MSMEG_1207|M.smegmatis_MC2_155 VIVVLHTVPSAPTAHQRAVLEQVAAL-AARVVVMSASASRRLCRDFRVDR
MAB_1019c|M.abscessus_ATCC_199 ESRFAG--AGPLRARAKYVVERLRYAGAERVIVLSDRFRDVLVEDYRIPT
. . :. :
MAV_4082|M.avium_104 SRTSVVPCGVDLDAFTPDGPVAARGARPRIVTVGRLVPRKGFDTIVRALP
TH_3188|M.thermoresistible__bu SRISVVPCGVDLDLFTPDGPQAPRGERRRIVTVGRFVPRKGFDTLVRALP
Mb3058|M.bovis_AF2122/97 AEITVIRNGIDAARW-PFAARRPRTGPAELLYVGRLEYEKGVHDAIAALP
Rv3032|M.tuberculosis_H37Rv AEITVIRNGIDAARW-PFAARRPRTGPAELLYVGRLEYEKGVHDAIAALP
MMAR_0474|M.marinum_M ERIAVVRNGLDEAPG-QTLSG-PRAATPRVVVLSRLVPHKQIEDALEAVA
MUL_1124|M.ulcerans_Agy99 ERIAVVRNGLDEAPG-QTLSG-PRAATPRVVVLSRLVPHKQIEDALEAVA
MLBr_02583|M.leprae_Br4923 ERIAVVRNGLDEAPA-QSLSG-TRGAAPRVVVLSRLVPHKQVEDALEAVA
Mvan_1042|M.vanbaalenii_PYR-1 RKVVTIPYGAVLPTG----PRAKRGSRPTILTCGLLGPGKGVERVIDVMS
Mflv_5224|M.gilvum_PYR-GCK HKIVVIPHGAVLPTQ----PRLKRGSRPVILSFGLLGPGKGVEHVIDAMP
MSMEG_1207|M.smegmatis_MC2_155 RKVITIPHGATLPCG----PVLKRGGRPTLLTWGLLGPGKGIERGIDAMS
MAB_1019c|M.abscessus_ATCC_199 DRIEVIPPGVDLDRFGVLPRLDAPTGRRTVLCVRRLEHRMGIHRLIESWP
. .: * :: : .. : .
MAV_4082|M.avium_104 RIPD----AELVIVGGPPAAELA--GDEQARRLQRLAGELGVADRVRLLG
TH_3188|M.thermoresistible__bu AIPN----AELVVVGGPRRSQLR--TEPEARRLCTLAEELGVADRVQLYG
Mb3058|M.bovis_AF2122/97 RLR-----RTHP---GTTLTIAG--EGTQQDWLIDQARKHRVLRATRFVG
Rv3032|M.tuberculosis_H37Rv RLR-----RTHP---GTTLTIAG--EGTQQDWLIDQARKHRVLRATRFVG
MMAR_0474|M.marinum_M QLR-----RRTPGSPNLHIDVVG--GGWWRQRLIDHVHRLGISDAVTFHG
MUL_1124|M.ulcerans_Agy99 QLR-----RRTPGSPNLHIDVVG--GGWWRQRLIDHVHRLGISDAVTFHG
MLBr_02583|M.leprae_Br4923 DLR-----QRLR-IPGLHLDIVG--GGWWRQRLVDHAHRLGISDAVTFHG
Mvan_1042|M.vanbaalenii_PYR-1 SLQSVPGHPRYVVAGRTHPKVLARDGEAYREARIEQARRLGVADSVTFED
Mflv_5224|M.gilvum_PYR-GCK SLLSVAGRPRYVVAGRTHPEVLARDGEAYREARVEQARAGGVADSVSFDD
MSMEG_1207|M.smegmatis_MC2_155 VLRRLNGMPRYLIAGLTHPKVLAAEGEAYRDACVERARSTGVSDAVYFDP
MAB_1019c|M.abscessus_ATCC_199 TVVS--------AHPDACLMIVG--TGTAEKDLRAQVAAAGLDDSVHFTG
: . : . :
MAV_4082|M.avium_104 -GVTRAQMPALLRSADVVACT--PWYEPFGIVPLEAMACGVPVVATA-VG
TH_3188|M.thermoresistible__bu -SVARADMPAVLRSADVVACT--PWYEPFGIVPLEAMACGVPVVAAA-VG
Mb3058|M.bovis_AF2122/97 -HLDHTELLALLHRADAAVLP--SHYEPFGLVALEAAAAGTPLVTSN-IG
Rv3032|M.tuberculosis_H37Rv -HLDHTELLALLHRADAAVLP--SHYEPFGLVALEAAAAGTPLVTSN-IG
MMAR_0474|M.marinum_M -HVDDVTKHHVLQSSWVQLLP--SRKEGWGLAVVEAAQHGVPTIGYRSSG
MUL_1124|M.ulcerans_Agy99 -HVDDVTKHHVLQSSWVQLLP--SRKEGWGLAVVEAAQHGVPTIGYRSSG
MLBr_02583|M.leprae_Br4923 -HVDDVTKHHVLQSSWLHLLP--SRKEGWGLAVVEAAQHNVPTIGYRSSG
Mvan_1042|M.vanbaalenii_PYR-1 RHLDRASLAALFQAAAVIVLPYDSTDQVTSGALVDAVASGRPVVATAFPH
Mflv_5224|M.gilvum_PYR-GCK RYLDRASLAALIQTAAVIVLPFDSKDQVTSGVLVDAVASGRPVVATAFPH
MSMEG_1207|M.smegmatis_MC2_155 GYQSASSLTTLVQAAKVVLLPYDSTEQVTSGVLVDAIAHGRPVVATAFPH
MAB_1019c|M.abscessus_ATCC_199 -RVDDLTLTQLYTLADFTVVP-SVALEGFGLIALESLATGRPAVVTD-CG
: : . : . ::: . * :
MAV_4082|M.avium_104 GIRDTVVDGATGRLVPPKDPDALGEAVAALLRDGRRGRTLGEAGRERARA
TH_3188|M.thermoresistible__bu GMLDTVVHDVTGRLVPPRRPAELADTINQLLRDSFLRQSLGAAGRDRARA
Mb3058|M.bovis_AF2122/97 GLGEAVINGQTGVSCAPRDVAGLAAAVRSVLDDPAAAQRRARAARQRLTS
Rv3032|M.tuberculosis_H37Rv GLGEAVINGQTGVSCAPRDVAGLAAAVRSVLDDPAAAQRRARAARQRLTS
MMAR_0474|M.marinum_M GLSDSIIDGVTGILVDS--HAELVDQLERLLADPVLRDQLGAKAQVR-SA
MUL_1124|M.ulcerans_Agy99 GLSDSIIDGVTGILVDS--HAELVDQLERLLADPVLRDQLGAKARVR-SA
MLBr_02583|M.leprae_Br4923 GLTDSIIDGVTGILVDN--RADLVDRLEELLTDPVLRDQLGAKAQAR-SL
Mvan_1042|M.vanbaalenii_PYR-1 AV--EVLRDGAGILVPHDDPEALSCALRRVLTQPRLAGSLAAEARQL-AP
Mflv_5224|M.gilvum_PYR-GCK AV--ELLCDGAGIVVPHDDPQALTAALRRVLSQPRLAGSMAAEARQL-AP
MSMEG_1207|M.smegmatis_MC2_155 AV--ELLSSGAGIVVDHDDPDALVSALRSVITQPRLAGAMAAEARRI-AP
MAB_1019c|M.abscessus_ATCC_199 GLPDAVRGLDSSLIVPADNAEALAARLVSALDGALPDPAQCRQHAES---
.: : :. * : :
MAV_4082|M.avium_104 RYSWDRVAADTERIYERLSPARRGVGTPSTRSG------
TH_3188|M.thermoresistible__bu RYSWDRVATDTQRIYDRLVPADYRSATASQMSSG-----
Mb3058|M.bovis_AF2122/97 DFDWQTVATATAQVYLAAKRGERQPQPRLPIVEHALPDR
Rv3032|M.tuberculosis_H37Rv DFDWQTVATATAQVYLAAKRGERQPQPRLPIVEHALPDR
MMAR_0474|M.marinum_M EFSWQESVDGMRSVLEAVPAGRFVSG----VV-------
MUL_1124|M.ulcerans_Agy99 EFSWQESVDGMRSVLEAVPAGRFVSG----VV-------
MLBr_02583|M.leprae_Br4923 EFSWRQSAGAMRTVLEAVQAGDYITG----VVSGGV---
Mvan_1042|M.vanbaalenii_PYR-1 AMAWPVVADTYLELAARLLTERQLRV-------------
Mflv_5224|M.gilvum_PYR-GCK TMAWPVVADAYAQLAARVVGERQMRM-------------
MSMEG_1207|M.smegmatis_MC2_155 SMAWSVVANAYQTVARELLTGRPRQR-------------
MAB_1019c|M.abscessus_ATCC_199 -FSWAAAAQRHHAVYAELSA-------------------
* . :