For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGSEWFSSLPGGLNRYFTDLFTALRTRGDIELSAAAFGVPSDGGSSWGGTGGSTRSRVLAAFNDRSGLQQ PGILDRHFCLYGPAAIGWGTRSRLVVHFHGPWAAESRFAGAGPLRARAKYVVERLRYAGAERVIVLSDRF RDVLVEDYRIPTDRIEVIPPGVDLDRFGVLPRLDAPTGRRTVLCVRRLEHRMGIHRLIESWPTVVSAHPD ACLMIVGTGTAEKDLRAQVAAAGLDDSVHFTGRVDDLTLTQLYTLADFTVVPSVALEGFGLIALESLATG RPAVVTDCGGLPDAVRGLDSSLIVPADNAEALAARLVSALDGALPDPAQCRQHAESFSWAAAAQRHHAVY AELSA*
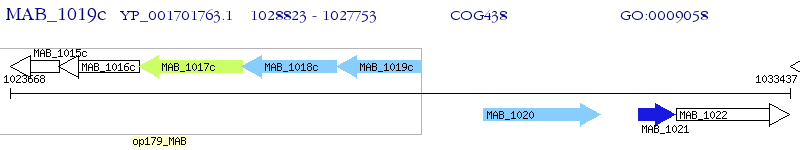
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1019c | - | - | 100% (356) | glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_3366 | - | 8e-20 | 28.90% (301) | glycosyltransferase |
| M. abscessus ATCC 19977 | MAB_1976 | - | 4e-18 | 31.36% (220) | hypothetical protein MAB_1976 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3058 | - | 1e-19 | 28.83% (385) | transferase |
| M. gilvum PYR-GCK | Mflv_4262 | - | 3e-21 | 32.78% (241) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv3032 | - | 1e-19 | 28.83% (385) | transferase |
| M. leprae Br4923 | MLBr_01715 | - | 1e-19 | 30.71% (241) | putative transferase |
| M. marinum M | MMAR_1681 | - | 1e-20 | 31.69% (243) | transferase |
| M. avium 104 | MAV_3879 | - | 6e-21 | 30.08% (246) | glycosyl transferase, group 1 family protein |
| M. smegmatis MC2 155 | MSMEG_2935 | - | 4e-19 | 31.42% (226) | phosphatidylinositol alpha-mannosyltransferase |
| M. thermoresistible (build 8) | TH_2191 | - | 1e-18 | 28.89% (315) | POSSIBLE TRANSFERASE |
| M. ulcerans Agy99 | MUL_1919 | - | 5e-21 | 31.69% (243) | transferase |
| M. vanbaalenii PYR-1 | Mvan_2096 | - | 2e-22 | 33.20% (241) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3058|M.bovis_AF2122/97 ------------------------MRILMVSWEYPPVVIGGLGRHVHHLS
Rv3032|M.tuberculosis_H37Rv ------------------------MRILMVSWEYPPVVIGGLGRHVHHLS
MLBr_01715|M.leprae_Br4923 MFIQRHAVVRGIFGTFAPAEGTTLMKILIVSWEYPPVVIGGLGRHVHHLS
MAV_3879|M.avium_104 ------------------------MKILMVSWEYPPVVIGGLGRHVHHLS
MMAR_1681|M.marinum_M ------------------------MKILMVSWEYPPVVIGGLGRHVHHLA
MUL_1919|M.ulcerans_Agy99 ------------------------MKILMVSWEYPPVVIGGLGRHVHHLA
TH_2191|M.thermoresistible__bu ------------------------MKILMVSWEYPPVVVGGLGRHVYHLS
Mflv_4262|M.gilvum_PYR-GCK ----------------------MKMKILLVSWEYPPVVIGGLGRHVHHLA
Mvan_2096|M.vanbaalenii_PYR-1 ----------------------MKTKILLVSWEYPPVVIGGLGRHVHHLA
MAB_1019c|M.abscessus_ATCC_199 ----------------------------MTGSEWFSSLPGGLNRYFTDLF
MSMEG_2935|M.smegmatis_MC2_155 -------------------------MRIGMVCPYSFDVPGGVQSHVLQLA
: : **: :. .*
Mb3058|M.bovis_AF2122/97 TALAAAGHDVVVLSRCPSGTDPSTHPSSDEVTEGVRVIAAAQDPHEFTFG
Rv3032|M.tuberculosis_H37Rv TALAAAGHDVVVLSRCPSGTDPSTHPSSDEVTEGVRVIAAAQDPHEFTFG
MLBr_01715|M.leprae_Br4923 TALAAAGHDVVVLSRRPSGTDPCTHPTSDEISEGVRVIAAAQDPHEFTFS
MAV_3879|M.avium_104 TALAAAGHDVVVLSRRPTGTDPRTHPSTDQISEGVRVIAAAQDPHEFTFG
MMAR_1681|M.marinum_M TALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVIAAAQDPHEFSFG
MUL_1919|M.ulcerans_Agy99 TALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVIAAAQDPHEFSFG
TH_2191|M.thermoresistible__bu TALAAAGHEVVVLSRRPTGTDPSTHPSTDEVRDGVRVLAAAQDPHEFDFG
Mflv_4262|M.gilvum_PYR-GCK TELVAAGHEVVVLSRRPTGTDPQTHPTTDETHEGVRVIAAAEDPHEFGFG
Mvan_2096|M.vanbaalenii_PYR-1 TELAQSGHEVVVLSRRPTATDPQTHPTTDEVHEGVRVIAAAEDPHEFTFG
MAB_1019c|M.abscessus_ATCC_199 TALRTRGDIELSAAAFGVPSDGGSSWGGTGGSTRSRVLAAFND-------
MSMEG_2935|M.smegmatis_MC2_155 EVLRDAGHEVSVLAPASPHVKLPDYVVSGGKAVPIPYNGSVAR-------
* *. : . .:
Mb3058|M.bovis_AF2122/97 NDMMAWTLAMGHAMIRAGLR----LKKLGTDRSWRPDVVHAHDWLVAHPA
Rv3032|M.tuberculosis_H37Rv NDMMAWTLAMGHAMIRAGLR----LKKLGTDRSWRPDVVHAHDWLVAHPA
MLBr_01715|M.leprae_Br4923 NDMMAWTLAMGHAMIRTGLS----LTRHSSDLPWRPDVVHAHDWLVAHPA
MAV_3879|M.avium_104 TDMMAWTLAMGHAMIRAGLPPKLHDKKPGGNRPWRPDVVHAHDWLVAHPA
MMAR_1681|M.marinum_M SDMMAWTLAMGHAMVRAGLT----LSGPGAEQPWQPDIVHAHDWLVAHPA
MUL_1919|M.ulcerans_Agy99 SDMMAWTLAMGHAMVRAGLT----LSGPGAEQPWQPDIVHAHDWLVAHPA
TH_2191|M.thermoresistible__bu RDMMAWTLAMGHSMVRAGLALG--LRGGTGGTPWQPDVVHAHDWLVAHPA
Mflv_4262|M.gilvum_PYR-GCK TDMMAWVLAMGHSMIRAGFG----------LDGWRPDIVHAHDWLVAHPA
Mvan_2096|M.vanbaalenii_PYR-1 TDMMAWTLAMGHAMIRAGLV----------LADWRPDIVHAHDWLVATPA
MAB_1019c|M.abscessus_ATCC_199 ---------------RSGLQ--------------QPGILDRHFCLYGPAA
MSMEG_2935|M.smegmatis_MC2_155 -------LRFGPATHRKVKK---------WIAEGDFDVLHIHEPNAPSLS
* .::. * :
Mb3058|M.bovis_AF2122/97 IALAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVHAVESWLVRESDSLI
Rv3032|M.tuberculosis_H37Rv IALAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVHAVESWLVRESDSLI
MLBr_01715|M.leprae_Br4923 ITLAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVHAVESWLVRESDSLI
MAV_3879|M.avium_104 IALAQFYDVPMVSTIHATEAGRHSGWVSGPISRQVHAIESWLVRESDSLI
MMAR_1681|M.marinum_M IALADYYDVPMVSTIHATEAGRHSGWVSGDLSRQVHAVESWLVRESDSLI
MUL_1919|M.ulcerans_Agy99 IALADYYDVPMVSTIHATEAGRHSGWVSGDLSRQVHAVESWLVRESDSLI
TH_2191|M.thermoresistible__bu IALAEFFDVPLVSTIHATEAGRHSGWVSGPISRQVHAVESWLVRDSDSLI
Mflv_4262|M.gilvum_PYR-GCK VALAQFFDVPLVSTIHATEAGRHSGWVSGSVSRQVHALESWLVRDSDSLI
Mvan_2096|M.vanbaalenii_PYR-1 VALAQFFDVPLVSTIHATEAGRHSGWVSGQVSRQVHALESWLVRDSDSLI
MAB_1019c|M.abscessus_ATCC_199 IGWGTRSRLVVHFHGPWAAESRFAGAGP-LRARAKYVVERLRYAGAERVI
MSMEG_2935|M.smegmatis_MC2_155 MLALQAAEGPIVATFHTSTT----------KSLTLSVFQGILRPYHEKII
: : : : ..: : :*
Mb3058|M.bovis_AF2122/97 TCSASMNDEITELFGPGLAEITVIRNGIDAARWPFAARRPR--TGPAELL
Rv3032|M.tuberculosis_H37Rv TCSASMNDEITELFGPGLAEITVIRNGIDAARWPFAARRPR--TGPAELL
MLBr_01715|M.leprae_Br4923 TCSASMCNEIIELFGPGLAEITVIRNGIDPARWPFAARRAR--TGPAELL
MAV_3879|M.avium_104 TCSASMADEITELFGPGLAQITVIRNGIETARWPFAPRRPR--TGPAELL
MMAR_1681|M.marinum_M TCSGSMSDEITELFGPGLAETAVIRNGIDAARWPFAVRKPR--TGPPELL
MUL_1919|M.ulcerans_Agy99 TCSGSMSDEITELFGPGLAETAVIRNGIDVARWPFAVRKPR--TGPPELL
TH_2191|M.thermoresistible__bu TCSASMSEEITELFGPGLSEIRVIRNGIDVAQWPFARRSPR--SGPPRLL
Mflv_4262|M.gilvum_PYR-GCK TCSASMRDEISALFGPELAEISVIPNGIDTDGWPFAARRAH--SGPAELL
Mvan_2096|M.vanbaalenii_PYR-1 TCSASMRDEITTLFGPELADITVIPNGIDTDGWPFAPHRPR--PGPAELL
MAB_1019c|M.abscessus_ATCC_199 VLSDRFRDVLVEDYRIPTDRIEVIPPGVDLDRFGVLPRLDAP-TGRRTVL
MSMEG_2935|M.smegmatis_MC2_155 GRIAVSDLARRWQMEALGSDAVEIPNGVDVASFADAPLLDGYPREGRTVL
* *:: : :*
Mb3058|M.bovis_AF2122/97 YVGRLEY-EKGVHDAIAALPRLRRTHPGTTLTIAGEGTQQDWLIDQARKH
Rv3032|M.tuberculosis_H37Rv YVGRLEY-EKGVHDAIAALPRLRRTHPGTTLTIAGEGTQQDWLIDQARKH
MLBr_01715|M.leprae_Br4923 YVGRLEY-EKGVHDVIAALPRIRRSYPGTTLTIAGEGTQQDWLVDQARKY
MAV_3879|M.avium_104 YLGRLEY-EKGVHDVIAALPRIRRTHPGTTLTIAGEGTQQGWLIEQARKH
MMAR_1681|M.marinum_M YVGRLEY-EKGVHNAIAALPRIRRAHPGTTLTIAGEGTQQDWLVEQARKH
MUL_1919|M.ulcerans_Agy99 YVGRLEY-EKGVHNAIAALPRIRRAHPGTTLTIAGEGTQQDWLVEQARKH
TH_2191|M.thermoresistible__bu YVGRLEY-EKGVQDAIAALPRIRRTHPGTTLTIAGTGTQHEWLVEQARKH
Mflv_4262|M.gilvum_PYR-GCK YFGRLEY-EKGVHDAIAALPKVRRTHPGTTLTIAGDGTQLGFLTEQARRH
Mvan_2096|M.vanbaalenii_PYR-1 YFGRLEY-EKGVHDLIAALPRIRRAHPGTTLTIAGDGTQLDFLVEQARKH
MAB_1019c|M.abscessus_ATCC_199 CVRRLEH-RMGIHRLIESWPTVVSAHPDACLMIVGTGTAEKDLRAQVAAA
MSMEG_2935|M.smegmatis_MC2_155 FLGRYDEPRKGMAVLLAALPKLVARFPDVEILIVGRG-DEDELREQAG--
. * : . *: : : * : .*.. : *.* * * *.
Mb3058|M.bovis_AF2122/97 RVLRATRFVGHLDHTELLALLHRADAAVLP-SHYEPFGLVALEAAAAGTP
Rv3032|M.tuberculosis_H37Rv RVLRATRFVGHLDHTELLALLHRADAAVLP-SHYEPFGLVALEAAAAGTP
MLBr_01715|M.leprae_Br4923 KVIKATRFVGHLNHNELLAALQRADAAVLP-SHYEPFGLVALEAAAAGTP
MAV_3879|M.avium_104 RVLKAIRFVGHLEHSELLAMLHRADAAVLP-SHYEPFGIVALEAAAAGTP
MMAR_1681|M.marinum_M RVLKATKFLGHLEHAELLTLLHRADAAVFP-SHYEPFGLVALEAAAAGTP
MUL_1919|M.ulcerans_Agy99 RVLKATKFLGHLEHAELLTLLHRADAAVFP-SHYEPFGLVALEAAAAGTP
TH_2191|M.thermoresistible__bu KVLKATRFVGQLGHEALLRQLHTADAAVLP-SHYEPFGIVALEAAATGAP
Mflv_4262|M.gilvum_PYR-GCK KVLKATKFVGRVDHSQLLTLLHRADVAVLP-SHYEPFGIVALEAAATGTP
Mvan_2096|M.vanbaalenii_PYR-1 KVVKATNFIGRVGHSELLRLLHRADAAVLP-SHYEPFGIVALEAAATGTP
MAB_1019c|M.abscessus_ATCC_199 GLDDSVHFTGRVDDLTLTQLYTLADFTVVPSVALEGFGLIALESLATGRP
MSMEG_2935|M.smegmatis_MC2_155 DLAGHLRFLGQVDDATKASAMRSADVYCAPHLGGESFGIVLVEAMAAGTA
: .* *:: . ** * * **:: :*: *:* .
Mb3058|M.bovis_AF2122/97 LVTSNIGGLGEAVINGQTGVSCAPRDVAGLAAAVRSVLDDPAAAQRRARA
Rv3032|M.tuberculosis_H37Rv LVTSNIGGLGEAVINGQTGVSCAPRDVAGLAAAVRSVLDDPAAAQRRARA
MLBr_01715|M.leprae_Br4923 LVTSNIGGLGEAVINGQTGVSCPPRDIAELAAMVCTVLEDPDAAQQRALA
MAV_3879|M.avium_104 LVTSNIGGLGEAVIDGQTGVSCPPRDVPALAAAIRTVLDDPEAAQRRACA
MMAR_1681|M.marinum_M LVTSNIGGLGEAVIDGVTGVSCPPRDVAALAAAVCGVLDDPAAAQQRARA
MUL_1919|M.ulcerans_Agy99 LVTSNIGGLGEAVIDGVTGVSCPPRDVAALAAAVCGVLDDPAAAQQRARA
TH_2191|M.thermoresistible__bu LVTSTAGGLGEAVIDGVTGKSFKPRDIAGLTAAVRAVLDDPEGAQQRAVA
Mflv_4262|M.gilvum_PYR-GCK LVTSTAGGLGEAVIDGVTGMSYPPSDVIALAAAIRAVLDAPAAAQERATA
Mvan_2096|M.vanbaalenii_PYR-1 LVTSTAGGLGEAVIDGVTGLSYPPGDVAALARAVRAVLDSPAAAQRRARA
MAB_1019c|M.abscessus_ATCC_199 AVVTDCGGLPDAVRGLDSSLIVPADNAEALAARLVSALDG----ALPDPA
MSMEG_2935|M.smegmatis_MC2_155 VVASDLDAFRRVLADGDAGRLVPVDDADGMAAALIGILEDDQLRAGYVAR
*.: ..: .: . :. : :: : *:
Mb3058|M.bovis_AF2122/97 ARQRLTSDFDWQTVATATAQVYLAAKRGERQPQPRLPIVEHALPDR--
Rv3032|M.tuberculosis_H37Rv ARQRLTSDFDWQTVATATAQVYLAAKRGERQPQPRLPIVEHALPDR--
MLBr_01715|M.leprae_Br4923 ARERLTSDFDWQTVAQQTAQVYLAAKRRERQPQPRLPIVEHALPDR--
MAV_3879|M.avium_104 ARERLTSDFDWHTVADKTAQVYLSAKRAERQPQARLPIVEHALPDR--
MMAR_1681|M.marinum_M ARERLNSDFDWQTVAEQTAQVYLAAKRGERQPLPRLPIVEHALPDR--
MUL_1919|M.ulcerans_Agy99 ARERLNSNFDWQTVAEQTAQVYLAAKRGERQPLPRLPIVEHALPDR--
TH_2191|M.thermoresistible__bu ARERLTADFDWHSVAAETAQVYLAAKRREREPLPRRLIVEHALPDR--
Mflv_4262|M.gilvum_PYR-GCK ARARLTSDFAWHTVAAETAQVYLAAKRAERRPQPRRTIVERPLPDR--
Mvan_2096|M.vanbaalenii_PYR-1 ARERLTSDFSWHTVAAETAQVYLAAKRAVRRPQPRRPIEERPLPDR--
MAB_1019c|M.abscessus_ATCC_199 QCRQHAESFSWAAAAQRHHAVYAELSA---------------------
MSMEG_2935|M.smegmatis_MC2_155 ASERVHR-YDWSVVSAQIMRVYETVSGAGIKVQVSGAANRDETAGESV
: : * .: ** .