For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSDLRSVLLLCWRDTGHPQGGGSETYLQRIGAQLAASGIEVTLRTARYPGAPRWELVDGVRVNRAGGRYS VYVWALLAMVAARMGLGPLRRVRPAAVIDTQNGLPFLARLVYGRRVVVLVHHCHREQWPVAGPVLGRLGW FVESKLSPWLNRRNQYVTVSLPSARDLVALGVDNERIAVVRNGLDEAPGQTLSGPRAATPRVVVLSRLVP HKQIEDALEAVAQLRRRTPGSPNLHIDVVGGGWWRQRLIDHVHRLGISDAVTFHGHVDDVTKHHVLQSSW VQLLPSRKEGWGLAVVEAAQHGVPTIGYRSSGGLSDSIIDGVTGILVDSHAELVDQLERLLADPVLRDQL GAKAQVRSAEFSWQESVDGMRSVLEAVPAGRFVSGVV
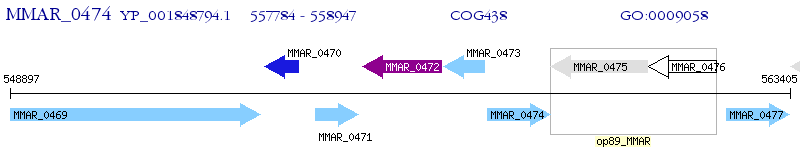
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0474 | - | - | 100% (387) | glycosyltransferase |
| M. marinum M | MMAR_1681 | - | 7e-19 | 26.36% (387) | transferase |
| M. marinum M | MMAR_3232 | - | 5e-15 | 25.91% (386) | glycosyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0230 | - | 0.0 | 83.46% (387) | hypothetical protein Mb0230 |
| M. gilvum PYR-GCK | Mflv_0426 | - | 1e-174 | 77.12% (389) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv0225 | - | 0.0 | 83.46% (387) | hypothetical protein Rv0225 |
| M. leprae Br4923 | MLBr_02583 | - | 0.0 | 82.86% (385) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_4480c | - | 1e-138 | 62.18% (386) | putative glycosyl transferase |
| M. avium 104 | MAV_4943 | - | 0.0 | 84.51% (381) | glycosyltransferase |
| M. smegmatis MC2 155 | MSMEG_0311 | - | 1e-167 | 73.70% (384) | glycosyltransferase |
| M. thermoresistible (build 8) | TH_2029 | - | 1e-167 | 75.07% (381) | POSSIBLE CONSERVED PROTEIN |
| M. ulcerans Agy99 | MUL_1124 | - | 0.0 | 99.22% (387) | glycosyltransferase |
| M. vanbaalenii PYR-1 | Mvan_0242 | - | 1e-173 | 76.86% (389) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0426|M.gilvum_PYR-GCK -MSDLP-----LRSVLLLCWRDTGHPQGGGSETYLQRIGAQLAASGTEVT
Mvan_0242|M.vanbaalenii_PYR-1 -MSDLP-----LRSVLLLCWRDTGHPQGGGSETYLQRIGAQLAESGTRVT
MMAR_0474|M.marinum_M -MSDLR-------SVLLLCWRDTGHPQGGGSETYLQRIGAQLAASGIEVT
MUL_1124|M.ulcerans_Agy99 -MSDLR-------SVLLLCWRDTGHPQGGGSETYLQRIGAQLAASGIEVT
Mb0230|M.bovis_AF2122/97 -MSALR-------SVLLLCWRDIGHPQGGGSEAYLQRIGAQLAASGIAVT
Rv0225|M.tuberculosis_H37Rv -MSALR-------SVLLLCWRDIGHPQGGGSEAYLQRIGAQLAASGIAVT
MLBr_02583|M.leprae_Br4923 -MFAARR---DLGSVLLLCWRDIGHPQGGGSETYLQRIGVQLAASGISVT
MAV_4943|M.avium_104 --------------MLLLCWRDTGHPQGGGSEAYLQRIGAQLAAAGIAVT
MSMEG_0311|M.smegmatis_MC2_155 -MSARPESAPHVRRVLLLCWRDTGHPQGGGSEAYVQRIGAYLAGRGVDVT
TH_2029|M.thermoresistible__bu --------------VLLLCWRDTGHPQGGGSETYLQRIGAQLAASGMRVT
MAB_4480c|M.abscessus_ATCC_199 MFSELSDQQDLPREVLMLCWRDTAHPQGGGSETYLQRIGAELAASGVKVT
:*:***** .********:*:****. ** * **
Mflv_0426|M.gilvum_PYR-GCK LRTARYPGAARDEVVDGVHVQRRGGSYTVYVWAGLAMAAARLGLGPLRRV
Mvan_0242|M.vanbaalenii_PYR-1 LRTARYPGAARREIVDGVQIQRRGGPYTVYVWAGLAMAAARIGLGPMRRI
MMAR_0474|M.marinum_M LRTARYPGAPRWELVDGVRVNRAGGRYSVYVWALLAMVAARMGLGPLRRV
MUL_1124|M.ulcerans_Agy99 LRTARYPGAPRWEVVDGVRVNRAGGRYSVYVWALLAMVAARMGLGPLQRV
Mb0230|M.bovis_AF2122/97 LRTARYPGAPRHELVDGVRISRAGGRYSVYLWALLAMAAARCGLGPLRRV
Rv0225|M.tuberculosis_H37Rv LRTARYPGAPRHELVDGVRISRAGGRYSVYLWALLAMAAARCGLGPLRRV
MLBr_02583|M.leprae_Br4923 LRTARYPGASRREVVDGVLISRAGGRYSVYLWALLAMAAARFGLGPLRRV
MAV_4943|M.avium_104 LRTARYPGAPRRDVVDGVRISRAGGRYSVYIWALLAMAVARLGLGPLRTV
MSMEG_0311|M.smegmatis_MC2_155 LRTARYPGSARTEVVDGVRISRGGGPYTVYIWAGLAMVLARIGLGPLRRA
TH_2029|M.thermoresistible__bu LRTARYRGAPRREIADGVHINRAGGRYTVYPRALLAMAAARLGLGPLRHV
MAB_4480c|M.abscessus_ATCC_199 LRCAAHPGAPRREVVDGVHISRAGGRFTVYPRALLSLLASRLGLGALRDV
** * : *:.* ::.*** :.* ** ::** * *:: :* ***.::
Mflv_0426|M.gilvum_PYR-GCK RPDVVVDTQNGLPFLSRLVHGRRAVVLVHHCHREQWPVAGPVMSRIGWFV
Mvan_0242|M.vanbaalenii_PYR-1 RPDIVVDTQNGLPFLSRLVHGRRTVVLVHHCHREQWPVAGPVMSRMGWFV
MMAR_0474|M.marinum_M RPAAVIDTQNGLPFLARLVYGRRVVVLVHHCHREQWPVAGPVLGRLGWFV
MUL_1124|M.ulcerans_Agy99 RPAAVIDTQNGLPFLARLVYGRRVVVLVHHCHREQWPVAGPVLGRLGWFV
Mb0230|M.bovis_AF2122/97 RPDVVVDTQNGWPFVARLLYGRRSLVLVHHCHREQWPVAGRMMGRLGWYV
Rv0225|M.tuberculosis_H37Rv RPDVVVDTQNGWPFVARLLYGRRSLVLVHHCHREQWPVAGRMMGRLGWYV
MLBr_02583|M.leprae_Br4923 RPDVVVDTQNGWPFLARLLYGSRVVVLVHHCHREQWPVAGPVIGRLGWYV
MAV_4943|M.avium_104 RPDVVVDTQNGLPFLARLIYGRRVVVLVHHCHREQWPVAGPVLGRLGWFV
MSMEG_0311|M.smegmatis_MC2_155 RPDVVIDTQNGLPFLARFAFGRRVAVLVHHCHRELWPVAGPVMGRIGWFV
TH_2029|M.thermoresistible__bu RPDVVVDTQNGVPFLARLVFGRRVAVLVHHCHRELWPVAGPVLGRVGWYV
MAB_4480c|M.abscessus_ATCC_199 RPDVVIDTQNGIPFFASTVAPAPVVVLVHHCHREQWPVAGPLVGRLGWWL
** *:***** **.: ********* ***** ::.*:**::
Mflv_0426|M.gilvum_PYR-GCK ESRLAPRLYRRNQYVTVSLPSARDLAVLGVPADHVAVVRNGVDAAP---G
Mvan_0242|M.vanbaalenii_PYR-1 ESRLSPRLHRRNQYVTVSLPSARDLALLGVQPERIAVVRNGVDAAP---E
MMAR_0474|M.marinum_M ESKLSPWLNRRNQYVTVSLPSARDLVALGVDNERIAVVRNGLDEAP---G
MUL_1124|M.ulcerans_Agy99 ESKLSPWLNRRNQYVTVSLPSARDLVALGVDNERIAVVRNGLDEAP---G
Mb0230|M.bovis_AF2122/97 ESMLSPRLHRRNQYVTVSLPSARDLIALGVDSERIAVVRNGLDEAP---S
Rv0225|M.tuberculosis_H37Rv ESMLSPRLHRRNQYVTVSLPSARDLIALGVDSERIAVVRNGLDEAP---S
MLBr_02583|M.leprae_Br4923 ESTLSPRLHRRNQYLTVSLPSARDLVALGVDHERIAVVRNGLDEAP---A
MAV_4943|M.avium_104 ESTLSPRLNRRNQYVTVSLPSARDLVALGVDGTRIAVVRNGLDEAP---A
MSMEG_0311|M.smegmatis_MC2_155 ESWLSPRLHRRNQYVTVSLPSARDLNELGVDSQRIAVVRNGLDEAP---A
TH_2029|M.thermoresistible__bu ESTISPRAHRGHQYVTVSLPSARDLMDLGVPPGHIAVVRNGLDEAP---P
MAB_4480c|M.abscessus_ATCC_199 ESRLSPRMHRRNQYLTVSQPSASDLVALGVEAGRIAVVRNGVDQTDPRIA
** ::* * :**:*** *** ** *** ::******:* :
Mflv_0426|M.gilvum_PYR-GCK ESLTVPRSATPRVVVLSRLVPHKQIEDALDVIAALRG--RIAD---VHLD
Mvan_0242|M.vanbaalenii_PYR-1 PTLAVPRSESPRVVVLSRLVPHKQIEDALDAVARLRT--RIPD---VHLD
MMAR_0474|M.marinum_M QTLSGPRAATPRVVVLSRLVPHKQIEDALEAVAQLRR--RTPGSPNLHID
MUL_1124|M.ulcerans_Agy99 QTLSGPRAATPRVVVLSRLVPHKQIEDALEAVAQLRR--RTPGSPNLHID
Mb0230|M.bovis_AF2122/97 PTLSGPRAPTPRVVVLSRLVPHKQIEDALAAVAELQP--RIPG---LHLD
Rv0225|M.tuberculosis_H37Rv PTLSGPRAPTPRVVVLSRLVPHKQIEDALAAVAELQP--RIPG---LHLD
MLBr_02583|M.leprae_Br4923 QSLSGTRGAAPRVVVLSRLVPHKQVEDALEAVADLRQRLRIPG---LHLD
MAV_4943|M.avium_104 PSLSGPRAAAPRVVVLSRLVPHKQIEDALEAVARLRP--RVPG---LHLD
MSMEG_0311|M.smegmatis_MC2_155 TTLKLPRSETPRLVVLSRLVPHKQIEDALEAVAQLRT--EMPD---VHLD
TH_2029|M.thermoresistible__bu DTLAVPRSATPRLVVLSRLVPHKQIEDALEAVARLRP--RIPE---LHLD
MAB_4480c|M.abscessus_ATCC_199 GDAMNVRSETPRAVVLSRLVPHKQIEHALTTLAVLRD--HVPD---VHLD
*. :** ***********:*.** .:* *: . . :*:*
Mflv_0426|M.gilvum_PYR-GCK VLGDGWWSQKLVEYAHRLGISDAVTFHGHVDDATKHVVLQRSWVHVLPSR
Mvan_0242|M.vanbaalenii_PYR-1 VLGDGWWAQRLVDHAARLGISDAVTFHGHVDDATKHAVLQRGWVHVLPSR
MMAR_0474|M.marinum_M VVGGGWWRQRLIDHVHRLGISDAVTFHGHVDDVTKHHVLQSSWVQLLPSR
MUL_1124|M.ulcerans_Agy99 VVGGGWWRQRLIDHVHRLGISDAVTFHGHVDDVTKHHVLQSSWVQLLPSR
Mb0230|M.bovis_AF2122/97 IVGGGWWRQRLVDHVHRLDIADAVTFHGHVDDVTKHHVLQSSWVHLLPSR
Rv0225|M.tuberculosis_H37Rv IVGGGWWRQRLVDHVHRLDIADAVTFHGHVDDVTKHHVLQSSWVHLLPSR
MLBr_02583|M.leprae_Br4923 IVGGGWWRQRLVDHAHRLGISDAVTFHGHVDDVTKHHVLQSSWLHLLPSR
MAV_4943|M.avium_104 IVGDGWWRQRLVEHVDRLGIGSAVTFHGHVDDVTKHHVLQGAWVHLLPSR
MSMEG_0311|M.smegmatis_MC2_155 ILGDGWWRERLVEHAELLGITDAVTFHGHVDEDTKHRVLQRSWVHVLPSR
TH_2029|M.thermoresistible__bu VLGDGWWADRLVDHARSLGIADAVTFHGHVDDETKHRVLQQSWVHVMPSR
MAB_4480c|M.abscessus_ATCC_199 VVGDGWWMDPLVEYAARLGVSDAVTFHGYLDEESKHQVLQRAWVHLMPSR
::*.*** : *:::. *.: .******::*: :** *** .*::::***
Mflv_0426|M.gilvum_PYR-GCK KEGWGLAVVEAAQHAVPTIGYRSSGGLTDSIVDGVTGVLVDGPADLADAL
Mvan_0242|M.vanbaalenii_PYR-1 KEGWGLSVVEAAQHGVPTIGYRSSGGLTDSVIDGVTGMLVDDFDELVDSL
MMAR_0474|M.marinum_M KEGWGLAVVEAAQHGVPTIGYRSSGGLSDSIIDGVTGILVDSHAELVDQL
MUL_1124|M.ulcerans_Agy99 KEGWGLAVVEAAQHGVPTIGYRSSGGLSDSIIDGVTGILVDSHAELVDQL
Mb0230|M.bovis_AF2122/97 KEGWGLAVIEAAQHGVPTIGYRSSGGLADSIVDGVTGILVDDRAELVAWL
Rv0225|M.tuberculosis_H37Rv KEGWGLAVIEAAQHGVPTIGYRSSGGLADSIVDGVTGILVDDRAELVAWL
MLBr_02583|M.leprae_Br4923 KEGWGLAVVEAAQHNVPTIGYRSSGGLTDSIIDGVTGILVDNRADLVDRL
MAV_4943|M.avium_104 KEGWGLAVVEAAQHRVPTIGYRSSGGLSDSIVDEVTGILVDTHAELVDRL
MSMEG_0311|M.smegmatis_MC2_155 KEGWGLAVTEAAQHAVPTIGYRSSGGLTDSVVDGVTGLLVDDRDGLVAGL
TH_2029|M.thermoresistible__bu KEGWGLAVTEAGQHAVPTVGYRSSGGLTDSIVDGVTGLLVDDRAELVGAL
MAB_4480c|M.abscessus_ATCC_199 KEGWGLAVTEAAQHGVPTVGYRSSGGLTDSVTDGITGLLCRDQDEFVRHT
******:* **.** ***:********:**: * :**:* :.
Mflv_0426|M.gilvum_PYR-GCK ERLLTDDVLREQLGAKAQVRSGEFSWKESADAMRTVLESVHAGRMTSGVL
Mvan_0242|M.vanbaalenii_PYR-1 ERVLTDDVLREQLGAKAQVRSGDFSWKQSADAMRAVLTAVRAGRPASGVI
MMAR_0474|M.marinum_M ERLLADPVLRDQLGAKAQVRSAEFSWQESVDGMRSVLEAVPAGRFVSGVV
MUL_1124|M.ulcerans_Agy99 ERLLADPVLRDQLGAKARVRSAEFSWQESVDGMRSVLEAVPAGRFVSGVV
Mb0230|M.bovis_AF2122/97 EQLLSDSVLRDQLGAKAQARSGEFSWRQSAEALRSVLEAVQASRFVSGVV
Rv0225|M.tuberculosis_H37Rv EQLLSDSVLRDQLGAKAQARSGEFSWRQSAEALRSVLEAVQASRFVSGVV
MLBr_02583|M.leprae_Br4923 EELLTDPVLRDQLGAKAQARSLEFSWRQSAGAMRTVLEAVQAGDYITGVV
MAV_4943|M.avium_104 EQLLADPVLRDQLGAKAHTRSGEFSWAQSADAMRGVLEAVQAGRTVSGLV
MSMEG_0311|M.smegmatis_MC2_155 RQLVSDPVLRTQLGSKAQTRSDEFSWAQSADAMRTVLESVRAGRYLSGLV
TH_2029|M.thermoresistible__bu ERLLTDPVLLAELGAKAQARSREFSWRQSAEAMRTVLQSVYDGRRVSGLI
MAB_4480c|M.abscessus_ATCC_199 ATLLADNKLRYRMGEAARARCADLSWRRSAAGVSAVLKAAMDGAPISGII
:::* * .:* *:.*. ::** .*. .: ** :. . :*::
Mflv_0426|M.gilvum_PYR-GCK --------------
Mvan_0242|M.vanbaalenii_PYR-1 --------------
MMAR_0474|M.marinum_M --------------
MUL_1124|M.ulcerans_Agy99 --------------
Mb0230|M.bovis_AF2122/97 --------------
Rv0225|M.tuberculosis_H37Rv --------------
MLBr_02583|M.leprae_Br4923 SGGV----------
MAV_4943|M.avium_104 SGRG----------
MSMEG_0311|M.smegmatis_MC2_155 --------------
TH_2029|M.thermoresistible__bu --------------
MAB_4480c|M.abscessus_ATCC_199 GAEPEIAPQVEIPA