For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKRDQLSDVFDLIEVAGMVSGGFSVRGDWFCRLEIKDPVKMIGVVCGRVRLATDGYGPIDLEPGDVAILN GCTQVVLKGGPGAAPATKLDVEQSGDAFVRVDGAECGEADVIIGGHVDVNPTGAALLAQALPRVGLVRAS ADEATNLHAILNRVLDEVTGNRIGSAFAVQQQGQLLLLEVLRAYLAQADDLPPGWLRLLTDERLRPAVTS MHDEPGKPWRLEELARAAAMSRTSFAERFRAVAGVPPLTYLNAWRMQLARHALRDSDTRVGPLAFRLGYT SESAFSNAFKREVGMSPLRYRASVQAV
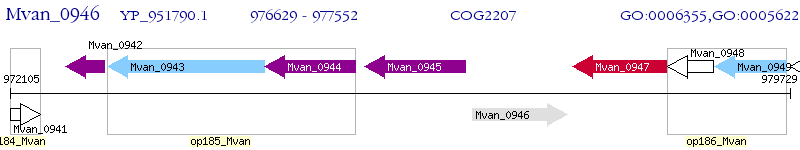
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0946 | - | - | 100% (307) | helix-turn-helix domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1013 | - | 4e-29 | 32.27% (251) | helix-turn-helix domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_0972 | - | 2e-10 | 32.00% (125) | AraC family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1966c | - | 2e-07 | 32.29% (96) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_5242 | - | 4e-26 | 31.42% (261) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | Rv1931c | - | 2e-07 | 32.29% (96) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0767c | - | 6e-24 | 32.28% (189) | AraC family transcriptional regulator |
| M. marinum M | MMAR_2855 | - | 7e-11 | 31.20% (125) | transcriptional regulatory protein |
| M. avium 104 | MAV_0221 | - | 5e-07 | 31.63% (98) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_4655 | - | 4e-11 | 37.00% (100) | transcriptional regulator, AraC family protein |
| M. thermoresistible (build 8) | TH_3120 | - | 1e-07 | 32.94% (85) | PUTATIVE transcriptional regulator, AraC family with |
| M. ulcerans Agy99 | MUL_2902 | - | 4e-11 | 31.20% (125) | transcriptional regulatory protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1966c|M.bovis_AF2122/97 ----MVIVGFPG--------------------------------------
Rv1931c|M.tuberculosis_H37Rv ----MVIVGFPG--------------------------------------
MMAR_2855|M.marinum_M MARKVVIVGFPGVQPLDVVGPFEVFAGASLLSEGGYDVALASIDERPVAT
MUL_2902|M.ulcerans_Agy99 MARKVVTVGFPGVQPLDVVGPFEVFAGASLLSEGGYDVALASIDERPVAT
TH_3120|M.thermoresistible__bu -VAALVLDGLAIFEFGVICEVFGVDRTDDGVPPVDFTVCGPRAG-QPVRT
Mvan_0946|M.vanbaalenii_PYR-1 MKRDQLSDVFDLIEVAGMVSG-GFSVRGDWFCRLEIKDPVKMIG------
MAB_0767c|M.abscessus_ATCC_199 MIHNPDYFADQPKSLSWPTGSPSIDAFSGILSAIGLRGESALRCS-----
Mflv_5242|M.gilvum_PYR-GCK MDVLMSLLRPQAVLSKVITGAGSWGVRKPRYGDPAFCLMLEG--------
MSMEG_4655|M.smegmatis_MC2_155 MQTERELLTEICERIDRHARGDMHTRIDGLLLSKAAGSSRPD--------
MAV_0221|M.avium_104 MYRPSPPLAEHIEYFGYWRG------------------------------
Mb1966c|M.bovis_AF2122/97 ---------------DPVDTVILPGGAGVDAARSEPALIDWVKAVSGTAR
Rv1931c|M.tuberculosis_H37Rv ---------------DPVDTVILPGGAGVDAARSEPALIDWVKAVSGTAR
MMAR_2855|M.marinum_M GIGLEFMTTRLPDPSAPVDTVVLPGGGGVDAARDNVALMDWVKAAAGTAR
MUL_2902|M.ulcerans_Agy99 GIGLEFMTTRLPDPSAPVDTVVLPGGGGVDAARDNVALMDWVKAAAGTAR
TH_3120|M.thermoresistible__bu SFGATLTPDSGLDGLAGADLVAIP---AIRQDDYLPEALDAVRAAADAGA
Mvan_0946|M.vanbaalenii_PYR-1 -----VVCGRVRLATDGYGPIDLEPGDVAILNGCTQVVLKGGPGAAPATK
MAB_0767c|M.abscessus_ATCC_199 ---AELPFDVSVPANERVLHIAETAGLRVYVDGNPVCVLAAGDMVLMARG
Mflv_5242|M.gilvum_PYR-GCK ---------SCVLDADGLDSIALSQGDFLLLPETPEFTLSAGPEATPSFS
MSMEG_4655|M.smegmatis_MC2_155 ------------YTVTEPLLVVMARGGKRIMVGDEVYEYRTGEMLIVTAS
MAV_0221|M.avium_104 ------------------DEALGVHTSRALPRGAVTAIIDVGGRTDLGFY
Mb1966c|M.bovis_AF2122/97 RVVTVCTGAFLAAEAGLLG--RTPSDDALG--LCRTFRPR-ISGRSGRCR
Rv1931c|M.tuberculosis_H37Rv RVVTVCTGAFLAAEAGLLG--RTPSDDALG--LCRTFRPR-ISGRSGRCR
MMAR_2855|M.marinum_M RVVTVCTGTFLAAEAGLLDGCRATTHWAFADRLAREFPAVDVDPDPIFIR
MUL_2902|M.ulcerans_Agy99 RVVTVCTGTFLAAEAGLLDGCRATTHWAFADRLAREFPAVDVDPDPIFIR
TH_3120|M.thermoresistible__bu IILTVCSGAFLAGAAGLLDGRPCTTHWGLTEELARRYPTARVDRNVLYVD
Mvan_0946|M.vanbaalenii_PYR-1 LDVEQSGDAFVRVDGAECGEADVIIGGHVDVNPTGAALLAQALPRVGLVR
MAB_0767c|M.abscessus_ATCC_199 TAHRLSGAQPHGEPAGTLH----WVSGRFTAERKAADPLLSVLPPAIIIQ
Mflv_5242|M.gilvum_PYR-GCK ALDHSPHTRHGDASAPVTMR---MLGGYFRFDPANAALLVALLPPVIHVR
MSMEG_4655|M.smegmatis_MC2_155 LPVTGHYLETDQPSLGMGLELRPAALAELALRAPARPALRTGSAGSAIAV
MAV_0221|M.avium_104 ASDARTPLTVPPAFAAGAG----ATAYVVRVAPAHTVMTIHFRPAGALAF
.
Mb1966c|M.bovis_AF2122/97 PDLHAQFAEGVDRGWSHRRHRPRAGTGRRRPRHRDCPDGCPLARPVSAPT
Rv1931c|M.tuberculosis_H37Rv PDLHAQFAEGVDRGWSHRRHRPRAGTGRRRPRHRDCPDGCPLARPVSAPT
MMAR_2855|M.marinum_M SSPKVWTAAGVTAGIDLALALVEGDHGTEIAQTVARWLVLYLRRPGGQ-T
MUL_2902|M.ulcerans_Agy99 SSPKVWTAAGVTAGIDLALALVEGDHGTEIAQTVARWLVLYLRRPCGQ-T
TH_3120|M.thermoresistible__bu -DGDLITSAGTAAGIDACLHLVRRELGSEVTNTIARHMVVPPQRDGGQRQ
Mvan_0946|M.vanbaalenii_PYR-1 ASADEATNLHAILNRVLDEVTGNRIGSAFAVQQQGQLLLLEVLRAYLAQA
MAB_0767c|M.abscessus_ATCC_199 AARPGHEWLPLSLQLILAEILTPSPGSWVMISRILDLLFIHAVREWSS-S
Mflv_5242|M.gilvum_PYR-GCK GGDPDAPRLTRLVELIAEEADADGACRDAILQRLVEVLLIEAARVQSAPD
MSMEG_4655|M.smegmatis_MC2_155 GNADAEMLDAAARLLRLLDRPDDAPVLAPLVEQEILWRLLTGPHGDTVRQ
MAV_0221|M.avium_104 LGSPLSDLEDALVGLEDLWGRDAALLREQLIDAGSPPRRVALLQAFLVRR
:
Mb1966c|M.bovis_AF2122/97 RWADPVRGSGVDATRQTDLDPPGAGGHRGRAGGAHRIGELAQRAAMSPRH
Rv1931c|M.tuberculosis_H37Rv RWADPVRGSGVDATRQTDLDPPGAGGHRGRAGGAHRIGELAQRAAMSPRH
MMAR_2855|M.marinum_M QFAAPVWMP--RAKRPSIRDVQEA--IEAQPGGPHSIEELAHRAAMSPRH
MUL_2902|M.ulcerans_Agy99 QFAAPVWMP--RAKRPSIRDVQEA--IEAQPGGPHSIEELAHRAAMSPRH
TH_3120|M.thermoresistible__bu FIDQPIPVR------HSEGFAPHLDWILSNLDKTHTVASLARRANMSPRT
Mvan_0946|M.vanbaalenii_PYR-1 DDLPPGWLR----LLTDERLRPAVTSMHDEPGKPWRLEELARAAAMSRTS
MAB_0767c|M.abscessus_ATCC_199 SHAEPGWLT----AAMDPRLAPVLTAIHNNLEHPWSIAELAQHARQSRSA
Mflv_5242|M.gilvum_PYR-GCK LAGPHRGLIT---GLADPVLAPALRALHADIAHGWTVEGLARTASVSRAV
MSMEG_4655|M.smegmatis_MC2_155 IALADSALT---------YINRAITWMRDNYASPVRIEDLARMAGMSTSV
MAV_0221|M.avium_104 MRRNAVWPP--------ARLAPVLR--GADLDPSMRVSKAQELSGLSRKR
: . : *
Mb1966c|M.bovis_AF2122/97 FTRVFSDEVGEAPGRYVERIRTEAARRQLEETHDTVVAIAARCGFGTAET
Rv1931c|M.tuberculosis_H37Rv FTRVFSDEVGEAPGRYVERIRTEAARRQLEETHDTVVAIAARCGFGTAET
MMAR_2855|M.marinum_M FTRVFTGEVGEAPRQYVERVRTEAARRQLEETEDTVVAIAARCGFGTAET
MUL_2902|M.ulcerans_Agy99 FTRVFTGEVGEAPGRYVERVRTEAARRQLEETEDTVVAIAARCGFGTAET
TH_3120|M.thermoresistible__bu FARRFVEETGRTPMQWVTDQRVLYARRLLEETDLDIHRIAARCGFGSTTL
Mvan_0946|M.vanbaalenii_PYR-1 FAERFRAVAGVPPLTYLNAWRMQLARHALRDSDTRVGPLAFRLGYTSESA
MAB_0767c|M.abscessus_ATCC_199 FTQRFTELLGQPPAAYIGERRLDRAAHLLRSTTDPVRQIGRAVGYTSDAA
Mflv_5242|M.gilvum_PYR-GCK FAQKFTRAMGLTPMQYLLEWRVAVAKDLMRSERLSIAQLAQRVGYQSATA
MSMEG_4655|M.smegmatis_MC2_155 FHRHFRAVTAMSPLQFQKRIRLQQARSLLVSQPGDVAAVAHLVGYDSASQ
MAV_0221|M.avium_104 FAALFRCEVGLSPKAYLRVLRLQAALRALDTP-ARGATIAADLGYFDQAH
* * . .* : * * : :. *:
Mb1966c|M.bovis_AF2122/97 MRRSFIRRVGISPDQYRKAFA-----------------
Rv1931c|M.tuberculosis_H37Rv MRRSFIRRVGISPDQYRKAFA-----------------
MMAR_2855|M.marinum_M MRRNFIRRVGISPDQYRKAFA-----------------
MUL_2902|M.ulcerans_Agy99 MRRNFIRRVGISPDQYRKAFA-----------------
TH_3120|M.thermoresistible__bu LRHHFRRVIGVTPSDYRRRFACGSTTSGSTESGAEVTD
Mvan_0946|M.vanbaalenii_PYR-1 FSNAFKREVGMSPLRYRASVQAV---------------
MAB_0767c|M.abscessus_ATCC_199 FSRAFQRRFGEPPLRWRTHHRSG---------------
Mflv_5242|M.gilvum_PYR-GCK FTTAFTRVAGAPPTEFARTLAPGGV-------------
MSMEG_4655|M.smegmatis_MC2_155 FTREYRRMFGAPPGQDAARLRAAHAPEPALLP------
MAV_0221|M.avium_104 FVREFRAFTGATPTQYARRRSSMPGHVELAR-------
: : * .*