For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IHNPDYFADQPKSLSWPTGSPSIDAFSGILSAIGLRGESALRCSAELPFDVSVPANERVLHIAETAGLRV YVDGNPVCVLAAGDMVLMARGTAHRLSGAQPHGEPAGTLHWVSGRFTAERKAADPLLSVLPPAIIIQAAR PGHEWLPLSLQLILAEILTPSPGSWVMISRILDLLFIHAVREWSSSSHAEPGWLTAAMDPRLAPVLTAIH NNLEHPWSIAELAQHARQSRSAFTQRFTELLGQPPAAYIGERRLDRAAHLLRSTTDPVRQIGRAVGYTSD AAFSRAFQRRFGEPPLRWRTHHRSG*
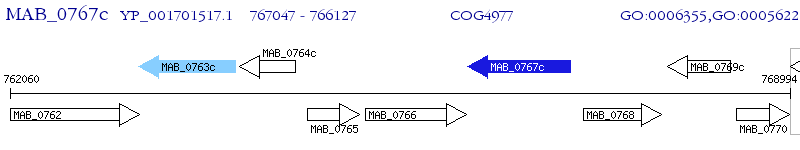
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0767c | - | - | 100% (306) | AraC family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1313 | - | 5e-12 | 36.45% (107) | AraC family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0942 | - | 3e-10 | 26.42% (193) | AraC family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1966c | - | 9e-07 | 34.57% (81) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_5242 | - | 1e-24 | 31.82% (242) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | Rv1931c | - | 9e-07 | 34.57% (81) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2855 | - | 1e-11 | 27.71% (231) | transcriptional regulatory protein |
| M. avium 104 | MAV_0193 | - | 1e-06 | 26.55% (113) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_1106 | - | 6e-13 | 28.43% (197) | transcriptional regulator, AraC family protein |
| M. thermoresistible (build 8) | TH_3120 | - | 8e-08 | 30.10% (103) | PUTATIVE transcriptional regulator, AraC family with |
| M. ulcerans Agy99 | MUL_2902 | - | 7e-12 | 27.71% (231) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_1013 | - | 9e-27 | 33.64% (220) | helix-turn-helix domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5242|M.gilvum_PYR-GCK ---------MDVLMSLLRPQAVLS--------------------------
Mvan_1013|M.vanbaalenii_PYR-1 ---------MEVLMSLLRPEAVLS--------------------------
Mb1966c|M.bovis_AF2122/97 -------------MVIVGFPG-----------------------------
Rv1931c|M.tuberculosis_H37Rv -------------MVIVGFPG-----------------------------
MMAR_2855|M.marinum_M ---------MARKVVIVGFPGVQPLDVVGPFEVFAGASLLSEG------G
MUL_2902|M.ulcerans_Agy99 ---------MARKVVTVGFPGVQPLDVVGPFEVFAGASLLSEG------G
MSMEG_1106|M.smegmatis_MC2_155 MSRVFDTEVVTRSVVILGYSGVQALDLVGPFDVFTGAALWLAGQGREQQA
TH_3120|M.thermoresistible__bu -----VAALVLDGLAIFEFGVICEVFGVDRTDDGVPPVDFTVCG------
MAB_0767c|M.abscessus_ATCC_199 --MIHNPDYFADQPKSLSWPTGSP--------------------------
MAV_0193|M.avium_104 --------------------------------------------------
Mflv_5242|M.gilvum_PYR-GCK -------------KVITGAGSWGVRKPRYGDPAFCLMLEGSCVLDADGLD
Mvan_1013|M.vanbaalenii_PYR-1 -------------KVITGGGRWSVRKPRYGDPAFCLMLEGLCVLDADGLD
Mb1966c|M.bovis_AF2122/97 ---------------------------DP---VDTVILPGGAGVDAARSE
Rv1931c|M.tuberculosis_H37Rv ---------------------------DP---VDTVILPGGAGVDAARSE
MMAR_2855|M.marinum_M YDVALASIDERPVATGIGLEFMTTRLPDPSAPVDTVVLPGGGGVDAARDN
MUL_2902|M.ulcerans_Agy99 YDVALASIDERPVATGIGLEFMTTRLPDPSAPVDTVVLPGGGGVDAARDN
MSMEG_1106|M.smegmatis_MC2_155 YTVTLASADGAPVTTLTGLEFVAAPPPDPCGDIDTVVIPGGIGADEARRN
TH_3120|M.thermoresistible__bu ------PRAGQPVRTSFGATLTPDSGLDGLAGADLVAIP---AIRQDDYL
MAB_0767c|M.abscessus_ATCC_199 -----------SIDAFSGILSAIGLRGESALRCSAELPFDVSVPANERVL
MAV_0193|M.avium_104 ----------------------MSEIGQHGLATSAQTLPPGARIARHRHP
Mflv_5242|M.gilvum_PYR-GCK SIALSQGDFLLLPETPEFTLSAGPEATPSFSALDHSPHTRHGDASAPVTM
Mvan_1013|M.vanbaalenii_PYR-1 PLELRQGDFLLLPQTPEFTLGAGGDVAPTFAGLDHSRHTRHGGESEPVTM
Mb1966c|M.bovis_AF2122/97 PALIDWVKAVSGTARRVVTVCTGAFLAAEAGLLGRTPSDDALGLCRTFRP
Rv1931c|M.tuberculosis_H37Rv PALIDWVKAVSGTARRVVTVCTGAFLAAEAGLLGRTPSDDALGLCRTFRP
MMAR_2855|M.marinum_M VALMDWVKAAAGTARRVVTVCTGTFLAAEAGLLDGCRATTHWAFADRLAR
MUL_2902|M.ulcerans_Agy99 VALMDWVKAAAGTARRVVTVCTGTFLAAEAGLLDGCRATTHWAFADRLAR
MSMEG_1106|M.smegmatis_MC2_155 REVVNWIKTAADHARRVVTVCTGAFLAAEAGLLDGCPATTHWASARRLAD
TH_3120|M.thermoresistible__bu PEALDAVRAAADAGAIILTVCSGAFLAGAAGLLDGRPCTTHWGLTEELAR
MAB_0767c|M.abscessus_ATCC_199 HIAETAGLRVYVDGNPVCVLAAGDMVLMARGTAHRLSGAQPHGEPAGTLH
MAV_0193|M.avium_104 LHQIVYP----STGAVSVTTPAGTWITPANRAIWIPAGCWHEHKFHGHTN
. :*
Mflv_5242|M.gilvum_PYR-GCK RMLGGYFRFDPA--NAALLVALLPPVIHVRGGDPDAPRLTRLVELIAEEA
Mvan_1013|M.vanbaalenii_PYR-1 RMLGGYFRFDAA--NAPLLVALLPPLILVRADQPGAQRLHRLVELITGEA
Mb1966c|M.bovis_AF2122/97 -------RISGR--SGR-----CRPDLHAQFAEGVDRGWSHRRHRPRAGT
Rv1931c|M.tuberculosis_H37Rv -------RISGR--SGR-----CRPDLHAQFAEGVDRGWSHRRHRPRAGT
MMAR_2855|M.marinum_M -------EFPAV--DVDPDPIFIRSSPKVWTAAGVTAGIDLALALVEGDH
MUL_2902|M.ulcerans_Agy99 -------EFPAV--DVDPDPIFIRSSPKVWTAAGVTAGIDLALALVEGDH
MSMEG_1106|M.smegmatis_MC2_155 -------EFPSV--TVDPEPIFMRSSERVWTAAGVTAGIDLALSLVEDDH
TH_3120|M.thermoresistible__bu -------RYPTA--RVDRNVLYVDDG-DLITSAGTAAGIDACLHLVRREL
MAB_0767c|M.abscessus_ATCC_199 WVSG---RFTAERKAADPLLSVLPPAIIIQAARPGHEWLPLSLQLILAEI
MAV_0193|M.avium_104 --------FHGV--------ALDPARYRRGPAAPAVLAVTPLMRELIIAC
Mflv_5242|M.gilvum_PYR-GCK DADGACRDAILQRLVEVLLIEAARVQSAPDLAGPHRGLITGLADPVLAPA
Mvan_1013|M.vanbaalenii_PYR-1 DADGACRDIILQRLVEVLLIEAMRVHTAADLGGAQRGLIAGLADPVLAPA
Mb1966c|M.bovis_AF2122/97 GRR---RPRHRDCPDGCPLARPVSAPTRWADPVRGSGVDATRQTDLDPPG
Rv1931c|M.tuberculosis_H37Rv GRR---RPRHRDCPDGCPLARPVSAPTRWADPVRGSGVDATRQTDLDPPG
MMAR_2855|M.marinum_M GTE---IAQTVARWLVLYLRRPGGQ-TQFAAPVWMP--RAKRPSIRDVQE
MUL_2902|M.ulcerans_Agy99 GTE---IAQTVARWLVLYLRRPCGQ-TQFAAPVWMP--RAKRPSIRDVQE
MSMEG_1106|M.smegmatis_MC2_155 GTD---VAQNVARWLVLYLRRPGGQ-TQFAAPVWMP--RAKRAPIRAVQE
TH_3120|M.thermoresistible__bu GSE---VTNTIARHMVVPPQRDGGQRQFIDQPIPVR------HSEGFAPH
MAB_0767c|M.abscessus_ATCC_199 LTPSPGSWVMISRILDLLFIHAVREWSSSSHAEPGW--LTAAMDPRLAPV
MAV_0193|M.avium_104 SRAR--DTDAGAHHRMLAVLHDQLQATSVAEPLWIP----TAVDGRLRAA
.
Mflv_5242|M.gilvum_PYR-GCK LRALHADIAHGWTVEGLARTASVSRAVFAQKFTRAMGLTPMQYLLEWRVA
Mvan_1013|M.vanbaalenii_PYR-1 LMELHADVTHGWTVEQLARVASVSRAVFAQRFTRTMGMPPMQYLLEWRVA
Mb1966c|M.bovis_AF2122/97 AGGHRGRAGGAHRIGELAQRAAMSPRHFTRVFSDEVGEAPGRYVERIRTE
Rv1931c|M.tuberculosis_H37Rv AGGHRGRAGGAHRIGELAQRAAMSPRHFTRVFSDEVGEAPGRYVERIRTE
MMAR_2855|M.marinum_M A--IEAQPGGPHSIEELAHRAAMSPRHFTRVFTGEVGEAPRQYVERVRTE
MUL_2902|M.ulcerans_Agy99 A--IEAQPGGPHSIEELAHRAAMSPRHFTRVFTGEVGEAPGRYVERVRTE
MSMEG_1106|M.smegmatis_MC2_155 A--IEADPGGAHSISDLAQRAAMSPRHFTRVFTEEVGQSPGVYVERIRID
TH_3120|M.thermoresistible__bu LDWILSNLDKTHTVASLARRANMSPRTFARRFVEETGRTPMQWVTDQRVL
MAB_0767c|M.abscessus_ATCC_199 LTAIHNNLEHPWSIAELAQHARQSRSAFTQRFTELLGQPPAAYIGERRLD
MAV_0193|M.avium_104 CALIADNLREPLTLRQIGERIGVGQRTLSRLFRDELAMTFPQWRTQVRLQ
: :.. . ::: * . . : *
Mflv_5242|M.gilvum_PYR-GCK VAKDLMRSERLSIAQLAQRVGYQSATAFTTAFTRVAGAPPTEFARTLAPG
Mvan_1013|M.vanbaalenii_PYR-1 LAKELLRTERPAIAQLAQRVGYQSATAFTTAFTRVAGCSPTEFARTSGYV
Mb1966c|M.bovis_AF2122/97 AARRQLEETHDTVVAIAARCGFGTAETMRRSFIRRVGISPDQYRKAFA--
Rv1931c|M.tuberculosis_H37Rv AARRQLEETHDTVVAIAARCGFGTAETMRRSFIRRVGISPDQYRKAFA--
MMAR_2855|M.marinum_M AARRQLEETEDTVVAIAARCGFGTAETMRRNFIRRVGISPDQYRKAFA--
MUL_2902|M.ulcerans_Agy99 AARRQLEETEDTVVAIAARCGFGTAETMRRNFIRRVGISPDQYRKAFA--
MSMEG_1106|M.smegmatis_MC2_155 AARRQLEESDDTVTAIAARCGFGTAETMRRNFIRRLGVSPDQYRKTSALT
TH_3120|M.thermoresistible__bu YARRLLEETDLDIHRIAARCGFGSTTLLRHHFRRVIGVTPSDYRRRFACG
MAB_0767c|M.abscessus_ATCC_199 RAAHLLRSTTDPVRQIGRAVGYTSDAAFSRAFQRRFGEPPLRWRTHHRSG
MAV_0193|M.avium_104 HALVLLAERR-DVTSVAAECGWATPSAFIDTYRRAFGHTPGRLAPRPS--
* : : :. *: : : : * * .*
Mflv_5242|M.gilvum_PYR-GCK GV-------------
Mvan_1013|M.vanbaalenii_PYR-1 GDTG-----------
Mb1966c|M.bovis_AF2122/97 ---------------
Rv1931c|M.tuberculosis_H37Rv ---------------
MMAR_2855|M.marinum_M ---------------
MUL_2902|M.ulcerans_Agy99 ---------------
MSMEG_1106|M.smegmatis_MC2_155 GR-------------
TH_3120|M.thermoresistible__bu STTSGSTESGAEVTD
MAB_0767c|M.abscessus_ATCC_199 ---------------
MAV_0193|M.avium_104 ---------------