For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRRWSALAVLTLAVLLIGIDGTVLALATPFISADLGATGTQILWIGDIYSFVLAALLISMGSLGDRIGHR KLLLCGATAFAVISAVTAYSTSPQMLILTRALLGMAGATLTPATLALIRGLFPDQRERSIAVGIWASAFS AGAALGPVVGGVLLEHFWWGSVFLINIPVMVILLVGGVLLLPEHRNPDPGPWDLPSVALSMVGMLAVVYA VKEGFAGLAHGVGPDVAVAGLVGAAALTMFVRRQLRLPAPLIDVRLFANPRFSGVVAANLLSVLGLSGLV FFLSQYFQLVHGYGPLKAGLAELPAAVTATVFGVLAGVAVRYVSHRAVLATGLALVGVAMAAMTTSLLVF TPVAPYLPLGVALFVVGVGLGLAFTVASDVILASVPRERAGAAAAVSETAYELGMALGIAMLGSIVTAVY RTVVIAPEIPSAAADLARDSLPGAIHAAHSLPADQQSTLLDAARASFTHGLASASAAGAVLLLTAAVAVW WLLRPGSPGAAEVGVLADPGVHRDPGRDARVDAPGGAELGDRNS*
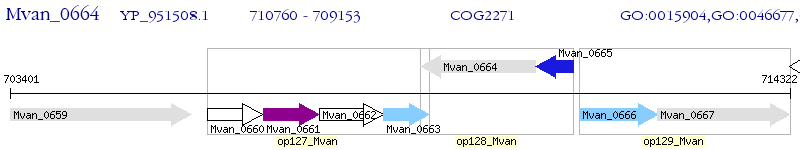
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0664 | - | - | 100% (535) | major facilitator superfamily transporter |
| M. vanbaalenii PYR-1 | Mvan_5021 | - | 4e-58 | 30.82% (425) | EmrB/QacA family drug resistance transporter |
| M. vanbaalenii PYR-1 | Mvan_0523 | - | 1e-57 | 31.60% (424) | EmrB/QacA family drug resistance transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2486 | - | 9e-51 | 29.71% (515) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_0241 | - | 0.0 | 81.87% (535) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv2459 | - | 9e-51 | 29.71% (515) | integral membrane transport protein |
| M. leprae Br4923 | MLBr_01562 | - | 5e-30 | 25.48% (416) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_2958 | - | 1e-100 | 41.38% (493) | putative transmembrane-transport protein |
| M. marinum M | MMAR_1146 | - | 9e-87 | 37.10% (504) | integral membrane transport protein |
| M. avium 104 | MAV_1387 | - | 2e-49 | 29.84% (516) | drug transporter |
| M. smegmatis MC2 155 | MSMEG_6225 | - | 1e-116 | 47.27% (495) | proton antiporter efflux pump |
| M. thermoresistible (build 8) | TH_3412 | - | 2e-97 | 40.16% (493) | PUTATIVE putative major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_0911 | - | 1e-86 | 36.90% (504) | integral membrane transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0664|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0241|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6225|M.smegmatis_MC2_155 -------------------------------------MSTCIEGTPSTTR
TH_3412|M.thermoresistible__bu -------------------------------------------MTQCAVR
MAB_2958|M.abscessus_ATCC_1997 -------------------------------MTSCKSTAPENATAPEGAR
MMAR_1146|M.marinum_M --------------------------------------------MRIQPV
MUL_0911|M.ulcerans_Agy99 --------------------------------------------MRIQPV
Mb2486|M.bovis_AF2122/97 --------------------------------------------------
Rv2459|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1387|M.avium_104 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Mvan_0664|M.vanbaalenii_PYR-1 -MTRRWSALAVLTLAVLLIGIDGTVLALATPFISADLGATGTQILWIGDI
Mflv_0241|M.gilvum_PYR-GCK -MSRRWLALGVLTLAVLLIGVDGTVLALATPFISADLGATGTELLWIGDI
MSMEG_6225|M.smegmatis_MC2_155 TPTRAWVALAVLALPVLLIAIDNTVLAFALPLIAEDFRPSATTQLWIVDV
TH_3412|M.thermoresistible__bu AGAREWAGLGVLAVPTVLLGLDVTVLYLVLPAVAADLAPSPVQTLWIMDA
MAB_2958|M.abscessus_ATCC_1997 VGRRTWFGLASLLLPVFLVSMDVSVLFLAMPRLSESLNPSAAEQLWILDV
MMAR_1146|M.marinum_M FRDHPIPALAVICLSVFVISVDATIVNVALPTLSRQLGADTAQLQWIVDA
MUL_0911|M.ulcerans_Agy99 FRDHPILALAVICLSVFVISVDATIVNVALPTLSRELGADTAQLQWIVDA
Mb2486|M.bovis_AF2122/97 MTPRQRLTVLATGLGIFMVFVDVNIVNVALPSIQKVFHTGEQGLQWAVAG
Rv2459|M.tuberculosis_H37Rv MTPRQRLTVLATGLGIFMVFVDVNIVNVALPSIQKVFHTGEQGLQWAVAG
MAV_1387|M.avium_104 -----------MMIGFFMIMVDSTIVAIANPTIMADLHIGYDTVVWVTSA
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
.: :* .: .. * : : *
Mvan_0664|M.vanbaalenii_PYR-1 YSFVLAALLISMGSLGDRIGHRKLLLCGATAFAVISAVTAYSTSPQMLIL
Mflv_0241|M.gilvum_PYR-GCK YSFVLAALLVTMGSLGDRVGHRKLLLCGATAFSAVSALTAYSTSPEMLIG
MSMEG_6225|M.smegmatis_MC2_155 YSLVLAALLVAMGSLGDRLGRRRLLLIGGAGFAVVSALAAFAPSAELLVG
TH_3412|M.thermoresistible__bu YGFFIAGFLITMGTLGDRIGRRRLLTIGVTAFAGASVLAALAPSAELLIV
MAB_2958|M.abscessus_ATCC_1997 YGFLLAGLLITMGNLGDRWGRRRLMLWGATLFAAASVLAAFAPTPLALIG
MMAR_1146|M.marinum_M YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLIA
MUL_0911|M.ulcerans_Agy99 YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLVA
Mb2486|M.bovis_AF2122/97 YSLGIAAVLMSCALLGDRYGRRRSFVFGVTLFVVSSIVCVLPVSLAVFTV
Rv2459|M.tuberculosis_H37Rv YSLGMAAVLMSCALLGDRYGRRRSFVFGVTLFVVSSIVCVLPVSLAVFTV
MAV_1387|M.avium_104 YLLGYAVVLLVAGRLGDRFGTKNLYLIGLAVFTVASVWCGLAGSAAMLIA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
* : . .:: . *.* * : * * * :
Mvan_0664|M.vanbaalenii_PYR-1 TRALLGMAGATLTPATLALIRGLFPDQRERSIAVGIWASAFSAGAALGPV
Mflv_0241|M.gilvum_PYR-GCK TRALLGMAGATLAPATLALIRGLFPDSRERSIAVGIWASAFSAGAALGPV
MSMEG_6225|M.smegmatis_MC2_155 ARALLGVFGAMLMPSTLSLIRNIFTDASARRLAIAIWASCFTAGSALGPI
TH_3412|M.thermoresistible__bu ARALLGVAGATLMPSTLSLISNMFVDAAQRALAIGVWATMFAVGMAAGPV
MAB_2958|M.abscessus_ATCC_1997 ARALMGVGGATLLPASLALIGVMFPNPRQKALAIGIWSAAFSFGAAVGPV
MMAR_1146|M.marinum_M ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
MUL_0911|M.ulcerans_Agy99 ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
Mb2486|M.bovis_AF2122/97 ARVIQGLGAAFISVLSLALLSHSFPNPRMKARAISNWMAIGMVGAASAPA
Rv2459|M.tuberculosis_H37Rv ARVIQGLGAAFISVLSLALLSHSFPNPRMKARAISNWMAIGMVGAASAPA
MAV_1387|M.avium_104 ARVVQGVGAGVMTPQTLSTITRIFPAER-RGVAVSVWSATAGAASLAGPL
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
:* *: .. *. : * : * . : : . .
Mvan_0664|M.vanbaalenii_PYR-1 VGGVLLEHFWWGSVFLINIPVMVILLVGGVLLLPEHRNPDPGPWDLPSVA
Mflv_0241|M.gilvum_PYR-GCK VGGVLLEHFWWGSVFLINIPVMVILLVGGLVLLPEHRDPDPGPWDLPSVA
MSMEG_6225|M.smegmatis_MC2_155 VGGALLEHFHWGAVFLVAVPILLPLLVLGPRLVPESRDPNPGPFDPVSIV
TH_3412|M.thermoresistible__bu VGGALVDRFWWGAAFLLAVPVAAAVLLG-ARLLPEYADPQAGRLDLRSVA
MAB_2958|M.abscessus_ATCC_1997 IGGVLLHHFWWGSVFLINVPVLIVLLLTADALIPEYRNPIRAPFDLVGVA
MMAR_1146|M.marinum_M TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLM
MUL_0911|M.ulcerans_Agy99 TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRVDVPGLM
Mb2486|M.bovis_AF2122/97 LGGLMVDGLGWRSVFLVNVPLGAIVWLLTLVGVDESQDPEPTQLDWVGQL
Rv2459|M.tuberculosis_H37Rv LGGLMVDGLGWRSVFLVNVPLGAIVWLLTLVGVDESQDPEPTQLDWVGQL
MAV_1387|M.avium_104 AGGVLVDGLGWQWIFFVNVPIGVLGLVLAYWLVPVLP-TQSHRFDLVGVG
MLBr_01562|M.leprae_Br4923 VGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKERMK-LDAAGAI
** : . * *:: :*: : * .
Mvan_0664|M.vanbaalenii_PYR-1 LSMVGMLAVVYAVKEGFAGLAHGVGPDVAVAGLVGAAALTMFVRRQ-LRL
Mflv_0241|M.gilvum_PYR-GCK LSMVGMLGIVYAIKEGFTGLVHGLGAEVVIAAAVGVAALTLFVRRQ-LRL
MSMEG_6225|M.smegmatis_MC2_155 LSFTTMLPIVWAVKT---AAHDGLSAAAAAAFAVGIVSGALFVRRQ-NRS
TH_3412|M.thermoresistible__bu LSLLAILAAIYAVKH---VAVHGVDGVAVAACALAAAAATVFVHRQ-RRL
MAB_2958|M.abscessus_ATCC_1997 LSMVGIMTLVYSVKA---MATHGFSAAVAITGAVGVLTLALFVRQQ-VRN
MMAR_1146|M.marinum_M LSAVGITALVYTIIEAP--NWGWTSTRASGGFIAAAIVLVGFALWE-RRS
MUL_0911|M.ulcerans_Agy99 LSAVGITALVYTIIEAP--NWGWTSTRASGGFIAAAIVLVGFALWE-RRS
Mb2486|M.bovis_AF2122/97 TLIPAVALIAYTIIEAP-RFDRQSAGFVAALLLAAGVLLWLFVRHE-HRA
Rv2459|M.tuberculosis_H37Rv TLIPAVALIAYTIIEAP-RFDRQSAGFVAALLLAAGVLLWLFVRHE-HRA
MAV_1387|M.avium_104 LSGVGMLLIVFGLQQGQ---AAHWQPWIWALIVAGVGFVTVFVFWQSVNV
MLBr_01562|M.leprae_Br4923 LATLACTAAVFAFSMGP--EKGWISLTTIGSGLVALVAAIAFAVVE-RTA
: . . *. :
Mvan_0664|M.vanbaalenii_PYR-1 PAPLIDVRLFANPRFSGVVAANLLSVLGLSGLVFFLSQYFQLVHGYGPLK
Mflv_0241|M.gilvum_PYR-GCK PHPLIDVRLFANPRFSGVVGANLFSVLGLSGLVFFLSQYFQLVHGYGPLK
MSMEG_6225|M.smegmatis_MC2_155 ATPMLDIGLFKVMPFTSSILANFLSIIGLIGFIFFISQHLQLVLGLSPLT
TH_3412|M.thermoresistible__bu DAPLLDVTLFADRAFTAALTVLLIGLIGVGGTMYLVTQFLQLVEGLTPVR
MAB_2958|M.abscessus_ATCC_1997 PHPLLALDLFRNRTFTVAIVGTVGAMATFGATTYLTGLYVQSVLGFDVLA
MMAR_1146|M.marinum_M SHPMLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQ
MUL_0911|M.ulcerans_Agy99 SHPMLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQ
Mb2486|M.bovis_AF2122/97 AFPLVDLKLFAEPLYRSVLIVYFVVMSCFFGTLMVITQHFQNVRDLSPLH
Rv2459|M.tuberculosis_H37Rv AFPLVDLKLFAEPLYRSVLIVYFVVMSCFFGTLMVITQHFQNVRDLSPLH
MAV_1387|M.avium_104 REPLIPLVIFSDRDFSLCNIGVAIISFAATAMMLPLTFYAQAVCGLSPTR
MLBr_01562|M.leprae_Br4923 ENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALR
*:: . :* . . * : .
Mvan_0664|M.vanbaalenii_PYR-1 AGLAELPAAVTATVFGVLAGVAVRYVSHRAVLATGLALVGVAMAAMTTSL
Mflv_0241|M.gilvum_PYR-GCK AGLAELPAAVTATVFGVLAGVAVRYFSHRSVLTTGLALVGTAMAALMASL
MSMEG_6225|M.smegmatis_MC2_155 AGLVTLPGAVVSMIAGLAVVKAAKRFAPDTLMVTGLVFVAVG----FLMI
TH_3412|M.thermoresistible__bu AGWWMGPPALAMFVAAIGAPLIARRVRPGVVMAATLGVSVLG-----YAV
MAB_2958|M.abscessus_ATCC_1997 AAFLGLPMAVVVAYFSMDAPRIERVLGERWTFVTSLLAMALG----NAVL
MMAR_1146|M.marinum_M TGVRLLPVALSIALAAILGPRLVERVGTTAVVAAGLAVFAAA----LAWA
MUL_0911|M.ulcerans_Agy99 TGVRLLPVAFSIALAAILGPRLVERVGTTAVVAAGLAVFAAA----LAWA
Mb2486|M.bovis_AF2122/97 AGLMMLPVPAGFGVASLLAGRAVNKWGPQLPVLTCLAAMFIG-----LAI
Rv2459|M.tuberculosis_H37Rv AGLMMLPVPAGFGVASLLAGRAVNKWGPQLPVLTCLAAMFIG-----LAI
MAV_1387|M.avium_104 SALLIAPMAIANGVFAPFVGKIVDRYHPRPVLGFGFSLLAIALT---WLT
MLBr_01562|M.leprae_Br4923 AGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLY--GWA
:. * .
Mvan_0664|M.vanbaalenii_PYR-1 LVFTPVAPYLPLGVALFVVGVGLGLAFTVASDVILASVPRERAGAAAAVS
Mflv_0241|M.gilvum_PYR-GCK LIFRPLTPYLPLGISLFVVGVGLGLAFTVASDVILASVPKERAGAAAAVS
MSMEG_6225|M.smegmatis_MC2_155 LLFRHNLTVAAIIASFVVLELGVGVSQTVSNDTIVASVPAAKSGAASAVS
TH_3412|M.thermoresistible__bu LASAGRGDGPAVVVAFAFVYLGLGAIAALGTDIVVGAAPAAKSGSAAAMS
MAB_2958|M.abscessus_ATCC_1997 LALGPHGPVAVYIAATVIVGAGAGVMFTFVSAVALGAAPTERAGQATGIS
MMAR_1146|M.marinum_M STADATTPYTQIALQMLLLGGGLGLTTAPATEAIMGSLPADRAGVGSAVN
MUL_0911|M.ulcerans_Agy99 STADATTPYTQITLQMLLLGGGLGLTTAPATEAIMGSLPADRAGVGSAVN
Mb2486|M.bovis_AF2122/97 FAISMDHAHPVALVGLTIFGAGAGGCATPLLHLGMTKVDDGRAGMAAGML
Rv2459|M.tuberculosis_H37Rv FAISMDHAHPVALVGLTIFGAGAGGCATPLLHLGMTKVDDGRAGMAAGML
MAV_1387|M.avium_104 FEMSPATPIWRLVLPFFAMGVGMAFVWSPLTATATRNLSAQLAGAGSAVY
MLBr_01562|M.leprae_Br4923 FMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIA
* . * :.:
Mvan_0664|M.vanbaalenii_PYR-1 ETAYELGMALGIAMLGSIVTAVYRT-VVIAPEIP----------------
Mflv_0241|M.gilvum_PYR-GCK ETAYELGMALGIAVLGSILTAVYRT-VTIPAGVP----------------
MSMEG_6225|M.smegmatis_MC2_155 ETAYELGAVVGTATLGTIFTAFYRSNVDVPAGLT----------------
TH_3412|M.thermoresistible__bu ETVQELGIAVGVALLGSVTAAVYRSRMTGPEGVA----------------
MAB_2958|M.abscessus_ATCC_1997 EMSFELGTAFGLALFGALASAVFAARTHVH--------------------
MMAR_1146|M.marinum_M DTTRELGGTLGVAIAGSVFASVYSGQLGSASTLAT---------------
MUL_0911|M.ulcerans_Agy99 DTTRELGGTLGVAIAGSVFASVYSGQLGSASALAT---------------
Mb2486|M.bovis_AF2122/97 NLQRSLGGIFGVAFLGTIVAAWLGAALPNTMADEIPDPI-----------
Rv2459|M.tuberculosis_H37Rv NLQRSLGGIFGVAFLGTIVAAWLGAALPNTMADEIPDPI-----------
MAV_1387|M.avium_104 NSVRQLGAVLGSAGMAAFMTWRIGAEMPGQPGGGGEDSAGPVLPEFLRGP
MLBr_01562|M.leprae_Br4923 LMLQSLGGPLVLAVIQAVITSRTLYLGGTTG-------------------
.** . * :. :
Mvan_0664|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0241|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6225|M.smegmatis_MC2_155 --------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
MAB_2958|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------------------------
Mb2486|M.bovis_AF2122/97 --------------------------------------------------
Rv2459|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1387|M.avium_104 FAAAMSQSVLLPAFIALFGIVAALFLVGFRPWAHRDGGTDAFGPDDYGGD
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mvan_0664|M.vanbaalenii_PYR-1 -------------------SAAADLARDSLPGAIHAAHSLPADQQSTLLD
Mflv_0241|M.gilvum_PYR-GCK -------------------ADAAAQARDSLPKAIDAARGLPDAAQSALLE
MSMEG_6225|M.smegmatis_MC2_155 -------------------PEQTGAAAESIGGAAAVAADLPAATATQLLD
TH_3412|M.thermoresistible__bu -------------------EGVGGPDAERLAESLPAAVSVADRVPGEAVR
MAB_2958|M.abscessus_ATCC_1997 ---------------------------ETLGQAMANAKELGGDAGAAAAE
MMAR_1146|M.marinum_M -----------------LPSDVRSAMKGSMAVAHKVIDQLPAGPAAGVRD
MUL_0911|M.ulcerans_Agy99 -----------------LPSDVRSAMKGSMAVAHKVIDQLPAGPAAGVRD
Mb2486|M.bovis_AF2122/97 ------------ARAIVVDVIVDSANPHAHAAFIGPGHRITAAQEDEIVL
Rv2459|M.tuberculosis_H37Rv ------------ARAIVVDVIVDSANPHAHAAFIGPGHRITAAQEDEIVL
MAV_1387|M.avium_104 YHDDYDDDDAYVELILVREPEPEAQQRAGQPRRPQPTPAPPADVRRRDPV
MLBr_01562|M.leprae_Br4923 ---------------------------PVKFMNDAQLRALDHGYTYGLLW
Mvan_0664|M.vanbaalenii_PYR-1 AARASFTHGLASASAAGAVLLLTAAVAVWWLLRPGSPGAAEVGVLADPGV
Mflv_0241|M.gilvum_PYR-GCK SAKESFTLGLSVSAGVGAALLLTSAVAVWWLLR--SPGPAEVGVLPDPGV
MSMEG_6225|M.smegmatis_MC2_155 SARAAFDSGIAPTAVIAAMLVLAAAAVVGVAFRR----------------
TH_3412|M.thermoresistible__bu LAQDAFAAGVNIAALIGGAAVVVVAVVCATALRHLRPLD-----------
MAB_2958|M.abscessus_ATCC_1997 LARTGWADGIHAVASVSTLLLVAIAVAAAVVMRRTQD-------------
MMAR_1146|M.marinum_M AVNHAFLDGLQVATLVCAGIALAAAIVVGWLLPARAGQPTPAQTEPALAQ
MUL_0911|M.ulcerans_Agy99 AVNHAFLDGLQVATLVCAGIALAAAIVVGWLLPARAGQPTPAQTEPALAQ
Mb2486|M.bovis_AF2122/97 AADAVFVSGIKLALGGAAVLLTGAFVLGWTRFPRTPAS------------
Rv2459|M.tuberculosis_H37Rv AADAVFVSGIKLALGGAAVLLTGAFVLGWTRFPRTPAS------------
MAV_1387|M.avium_104 ESRRSVLDERPAQVQPIGFAHNGSHVDGGKRLRQVAVRRAPKPGPPADRF
MLBr_01562|M.leprae_Br4923 VAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL-------------
.
Mvan_0664|M.vanbaalenii_PYR-1 HRDPGRDARVDAPGGAELGDRNS----------------------
Mflv_0241|M.gilvum_PYR-GCK HHDARGDGGVDTAGGAELGNRHS----------------------
MSMEG_6225|M.smegmatis_MC2_155 ---------------------------------------------
TH_3412|M.thermoresistible__bu ---------------------------------------------
MAB_2958|M.abscessus_ATCC_1997 ---------------------------------------------
MMAR_1146|M.marinum_M A--------------------------------------------
MUL_0911|M.ulcerans_Agy99 V--------------------------------------------
Mb2486|M.bovis_AF2122/97 ---------------------------------------------
Rv2459|M.tuberculosis_H37Rv ---------------------------------------------
MAV_1387|M.avium_104 TRPPRRHHPGPAGHHLGEGESLRGQHHRPDPDDDPTGYGRHSSGN
MLBr_01562|M.leprae_Br4923 ---------------------------------------------