For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQAERPLAADDHDRIVAAVHDEVARWGIDRFDVGAMAHRHGLDEPSIRRHWPEPESLLLHALALRPGDDG SLPDTGSLRTDLIALAQRMVALITSDEGRKLHGGHLIADTFVASMDVRRSAWRARAASLAGVFERARERG ELRAGVDHITALEMLFAPINMRALYTGEDIDDAYCEQIADMVYRAVSA
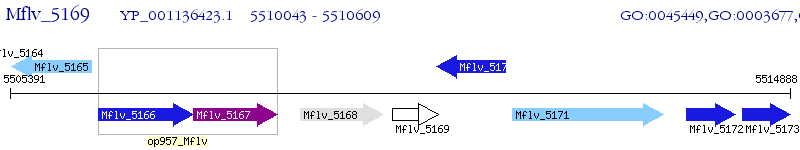
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5169 | - | - | 100% (188) | hypothetical protein Mflv_5169 |
| M. gilvum PYR-GCK | Mflv_4883 | - | 2e-27 | 37.63% (186) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3683 | - | 1e-11 | 31.79% (173) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2274c | - | 0.0001 | 33.33% (54) | transcriptional regulator |
| M. tuberculosis H37Rv | Rv2250c | - | 2e-05 | 29.55% (88) | transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0300c | - | 3e-20 | 31.03% (174) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0340 | - | 5e-05 | 27.74% (137) | TetR family transcriptional regulator |
| M. avium 104 | MAV_5165 | - | 2e-11 | 29.94% (177) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_3410 | - | 3e-17 | 33.33% (189) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3283 | - | 1e-09 | 28.09% (178) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_4780 | - | 2e-05 | 27.74% (137) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1163 | - | 6e-77 | 73.89% (180) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5169|M.gilvum_PYR-GCK ------MQAERPLAADDHDRIVAAVHDEVARWGIDRFDVGAMAHRHGLDE
Mvan_1163|M.vanbaalenii_PYR-1 ------MTRDHSGADDDRDRVIAAVHDELARWGIDRFDVGAMAHRHGLDV
MMAR_0340|M.marinum_M ----MVADFGRPRDPRIDGAVLRATVELLAETGYAGLLVSAIAKRAGTSK
MUL_4780|M.ulcerans_Agy99 ----MVADFGRPRDPRIDGAVLRATVELLAETGYAGLLVSAIAKRAGTSK
MAV_5165|M.avium_104 ------MDFGRPRDPRIDAAVLRATVELLAETGYPGLLVSAIAQRAGTSK
TH_3283|M.thermoresistible__bu MPGSGARSPGRPRDPALDDAIRAAARDVVVEHGYDATTLALIADRAGVSV
MSMEG_3410|M.smegmatis_MC2_155 --------MARPRDDSITEAVVDATRQLCGEIGYRNLTMEALAARAGTTK
Mb2274c|M.bovis_AF2122/97 ------MLSMSNDRADTGGRILRAAASCVVDYGVDRVTLAEIARRAGVSR
Rv2250c|M.tuberculosis_H37Rv ------MLSMSNDRADTGGRILRAAASCVVDYGVDRVTLAEIARRAGVSR
MAB_0300c|M.abscessus_ATCC_199 ---MSTEKKPQIRGVNLERAVLAATLAQIEAVGIEQTRIAEIAARAGVHE
: *. * : :* * *
Mflv_5169|M.gilvum_PYR-GCK PSIRRHWPEPESLLLHALALRPGDDGSLPDTGSLRTDLIALAQRMVALIT
Mvan_1163|M.vanbaalenii_PYR-1 EAIRRRWPDPESLVLDALAERPGDDGAVPDTGSLRTDLFHLAGRMAAIVT
MMAR_0340|M.marinum_M PAIYRRWPSKAHLVHEAVFP-IDAATAVPDTGSPVDDLREMVRRTTAVLS
MUL_4780|M.ulcerans_Agy99 PAIYRRWPSKAHLVHEAVFP-IDAATAVPDTGSPVDDLREMVRRTTAVLS
MAV_5165|M.avium_104 PAIYRRWPSKAHLVHEAVFP-VGADTAIPDTGSTADDLREMVRRTMVFLT
TH_3283|M.thermoresistible__bu PTIYRRWQHKHELIEEAVLH-LDTFELPEPTGDLEADLGTWVRLFLTVAM
MSMEG_3410|M.smegmatis_MC2_155 PAIRRRWPSLRHVVVDAMTR-DNVSMAVPDTGSTRSDLIVLVEELRVSMG
Mb2274c|M.bovis_AF2122/97 PTVYRRWPDTRSIMASMLTS--HIAAVLREVPLDGDDREALVKQIVAVAD
Rv2250c|M.tuberculosis_H37Rv PTVYRRWPDTRSIMASMLTS--HIADVLREVPLDGDDREALVKQIVAVAD
MAB_0300c|M.abscessus_ATCC_199 TSIYRRWKTLPRLLADALIIHIDADIPIPDTGSLREDLMSLARGLAQFLE
:: *:* :: : . * .
Mflv_5169|M.gilvum_PYR-GCK SDEGRKLHGGHLIADTFVAS--MDVRRSAWRARAASLAGVFERARERGEL
Mvan_1163|M.vanbaalenii_PYR-1 SEAGRKLHGGHLIGDVHVAS--VEIRRSAWRSRAADLGVVFERARQRGEM
MMAR_0340|M.marinum_M TPAAKAALPGLIGEMAADPTLHSALLVRFGEIFGHGLTTWLDNAAARGQV
MUL_4780|M.ulcerans_Agy99 TPAAKAAQPGLIGEMAADPTLHSALLVRFGEIFGHGLTTWLDNAAARGQV
MAV_5165|M.avium_104 TPAAKAALPGLIGEMAADPSLHSALLERFAGAIGGGLADWLAAAAARGEA
TH_3283|M.thermoresistible__bu EPAARAAVPGLMSAYSRNPDDYRRLMTRGELPVRAAIAAMLERAKAAGQA
MSMEG_3410|M.smegmatis_MC2_155 DVMLGRILPALVVDLSDDGDLRRRFLDSVWRPRRERADRILRRGVERGEL
Mb2274c|M.bovis_AF2122/97 RLRGDDLIMSVMHSELARVYITERLGTSQQVLIEGLAARLTVAQRSGSVR
Rv2250c|M.tuberculosis_H37Rv RLRGDDLIMSVMHSELARVYITERLGTSQQVLIEGLAARLTVAQRSGSVR
MAB_0300c|M.abscessus_ATCC_199 TPHGDALMRGTVQVS--DDPEVAEARREFWNRRLTAVEAIVQRALVRGEV
. : .
Mflv_5169|M.gilvum_PYR-GCK RAGVDHITALEMLFAPINMRALYTGEDID----DAYCEQIADMVYRAVSA
Mvan_1163|M.vanbaalenii_PYR-1 AEDIDHFTVLELLFAPINMRALYTGEPID----DRYCRTIADLVYRAVSP
MMAR_0340|M.marinum_M RADVTAPELAEAIAGIVLIGLLTRPGELEPGVGDAWIDRTTTLLLKGIRK
MUL_4780|M.ulcerans_Agy99 RADVTAPELAEAIAGIVLIGLLTRPGELEPGVGDAWIDRTTTLLLKGIRK
MAV_5165|M.avium_104 RPDVTAAELAETIAGVTLVALLTRPTELD----DAWVDRTTRLLLKGISA
TH_3283|M.thermoresistible__bu APDCDPDAVFELIRGATFLRALTHSDEDA----EAFCTRITEAVLRAVRS
MSMEG_3410|M.smegmatis_MC2_155 RADLDFELAQDLLAATVIYRFLFAHADLDP----ALSEHIVDYVLRGIAA
Mb2274c|M.bovis_AF2122/97 SGDARRLATMVLLIAQSTIQSADIVDSILDS--AALATELTHALNGYLC-
Rv2250c|M.tuberculosis_H37Rv SGDARRLATMVLLIAQSTIQSADIVDSILDS--AALATELTHALNGYLC-
MAB_0300c|M.abscessus_ATCC_199 RPGVDARTVVLTLGGYVHLWAAHLGQTAD----DLALDELVDLIIDGVSA
. : . . : :
Mflv_5169|M.gilvum_PYR-GCK ------
Mvan_1163|M.vanbaalenii_PYR-1 A-----
MMAR_0340|M.marinum_M GTPQ--
MUL_4780|M.ulcerans_Agy99 GTPQ--
MAV_5165|M.avium_104 ------
TH_3283|M.thermoresistible__bu ------
MSMEG_3410|M.smegmatis_MC2_155 GPTESS
Mb2274c|M.bovis_AF2122/97 ------
Rv2250c|M.tuberculosis_H37Rv ------
MAB_0300c|M.abscessus_ATCC_199 GR----