For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSDNDESRRAAAIEAALDEVQQWGVDRFRLEGVAHRAKLSPDYVRQIWGSETELIAEALLSYSQAMMELP DTGSLHGDLTGLALALADYLNEPVGRRISRMLVIDSKSQAVDRDTRARFWTVRRAIVEDIFRRAAERGEL RDDVRHIVALQLLTSPLHTFALYSEDPIPPEYCRLIADLVTRAIRAD
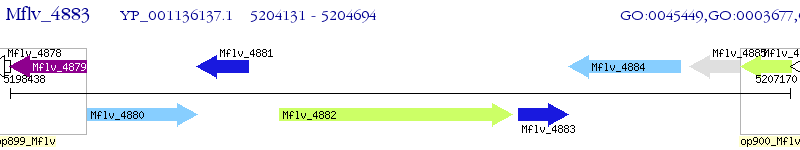
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4883 | - | - | 100% (187) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_5169 | - | 2e-27 | 37.63% (186) | hypothetical protein Mflv_5169 |
| M. gilvum PYR-GCK | Mflv_3683 | - | 4e-11 | 26.97% (178) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1581 | - | 2e-09 | 28.68% (136) | regulatory protein |
| M. tuberculosis H37Rv | Rv1556 | - | 2e-09 | 28.68% (136) | regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0300c | - | 5e-16 | 32.00% (175) | TetR family transcriptional regulator |
| M. marinum M | MMAR_1695 | - | 4e-11 | 25.15% (171) | regulatory protein |
| M. avium 104 | MAV_3866 | - | 3e-11 | 30.22% (139) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_3410 | - | 3e-14 | 30.65% (186) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3283 | - | 3e-07 | 27.59% (174) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_1934 | - | 2e-11 | 25.15% (171) | regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_1545 | - | 2e-63 | 61.62% (185) | hypothetical protein Mvan_1545 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1581|M.bovis_AF2122/97 MVGAVTQIADRPTDPSPWSPRETELLAVTLRLLQEHGYDRLTVDAVAASA
Rv1556|M.tuberculosis_H37Rv MVGAVTQIADRPTDPSPWSPRETELLAVTLRLLQEHGYDRLTVDAVAASA
MMAR_1695|M.marinum_M ----MTQTADRSAQVSPWSPREAELLAVTLQLLQEHGYDRLTIDAVANTA
MUL_1934|M.ulcerans_Agy99 -MGAMTQTADRSAQVSPWSPREAELLAVTLQLLQEHGYDRLTIDAVANTA
MAV_3866|M.avium_104 ----MTQTADRCEQASPWSPREAEILAVTLRLLQEHGYDQLTVDAVAGAA
Mflv_4883|M.gilvum_PYR-GCK ----MSDNDE---------SRRAAAIEAALDEVQQWGVDRFRLEGVAHRA
Mvan_1545|M.vanbaalenii_PYR-1 ----MSDVDA---------SQRATAIEAALAELQQWGVDRFSIEGVAQRS
MAB_0300c|M.abscessus_ATCC_199 ----MSTEKKPQIRG---VNLERAVLAATLAQIEAVGIEQTRIAEIAARA
MSMEG_3410|M.smegmatis_MC2_155 ----MARPRDD--------SITEAVVDATRQLCGEIGYRNLTMEALAARA
TH_3283|M.thermoresistible__bu ----MPGSGARSPGRPRDPALDDAIRAAARDVVVEHGYDATTLALIADRA
:. .: * : :* :
Mb1581|M.bovis_AF2122/97 RASKATVYRRWPSKAELVLAAFIEGIRQVAVPPNTGNLRDDLLRLGELIC
Rv1556|M.tuberculosis_H37Rv RASKATVYRRWPSKAELVLAAFIEGIRQVAVPPNTGNLRDDLLRLGELIC
MMAR_1695|M.marinum_M RASKATVYRRWPSKAELVLAAFIEGVRQVAVAPNTGSLRDDLVKIGEGIC
MUL_1934|M.ulcerans_Agy99 RASKATVYRRWPSKAELVLAAFIEGVRQVAVAPNTGSLRDDLVKIGEGIC
MAV_3866|M.avium_104 HASKATVYRRWPSKAELVLAAFIEGVRQVAVPPNTGTLRGDLLALGETVC
Mflv_4883|M.gilvum_PYR-GCK KLSPDYVRQIWGSETELIAEALLSYSQAMMELPDTGSLHGDLTGLALALA
Mvan_1545|M.vanbaalenii_PYR-1 RLDAEYLHEQWTSERQLIIDALLSYSDTIISVPDTGSLQGDLTELAMAIA
MAB_0300c|M.abscessus_ATCC_199 GVHETSIYRRWKTLPRLLADALIIHIDADIPIPDTGSLREDLMSLARGLA
MSMEG_3410|M.smegmatis_MC2_155 GTTKPAIRRRWPSLRHVVVDAMTR-DNVSMAVPDTGSTRSDLIVLVEELR
TH_3283|M.thermoresistible__bu GVSVPTIYRRWQHKHELIEEAVLHLDTFELPEP-TGDLEADLGTWVRLFL
: . * .:: *. * ** . ** .
Mb1581|M.bovis_AF2122/97 REVGQHAS--TIRAVLVEVSRNPALNDVLQHQFVDHRKALIQYILQQAVD
Rv1556|M.tuberculosis_H37Rv REVGQHAS--TIRAVLVEVSRNPALNDVLQHQFVDHRKALIQYILQQAVD
MMAR_1695|M.marinum_M GQAHQQAS--TIRAVLVEVSRHPALSDALQRQFVDQRKALIQQVLQQAVD
MUL_1934|M.ulcerans_Agy99 GQAHQQAS--TIRAVLVEVSRHPALSDALQRQFVDQRKALIQQVLQQAVD
MAV_3866|M.avium_104 EQVGHHAS--TIRAVMFEVSRHPALNDALQHQFLDQRRALIEHVLHQAVD
Mflv_4883|M.gilvum_PYR-GCK DYLNEPVGR-RISRMLVIDSKSQAVDRDTRARFWTVRRAIVEDIFRRAAE
Mvan_1545|M.vanbaalenii_PYR-1 AYVNQPMGR-RIARMMVVDTKSHTADYGTRSQFWTMRTEAIEVIFSRAAE
MAB_0300c|M.abscessus_ATCC_199 QFLETPHGD-ALMRGTVQVSDDPEVAEARR-EFWNRRLTAVEAIVQRALV
MSMEG_3410|M.smegmatis_MC2_155 VSMGDVMLGRILPALVVDLSDDGDLRRRFLDSVWRPRRERADRILRRGVE
TH_3283|M.thermoresistible__bu TVAMEPAARAAVPGLMSAYSRNPDDYRRLMTRGELPVRAAIAAMLERAKA
: : :. :.
Mb1581|M.bovis_AF2122/97 RGEISSAAISDELWDLLPGYLIFRSIIPNRPPTQDTVQALVDDVILPSLT
Rv1556|M.tuberculosis_H37Rv RGEISSAAISDELWDLLPGYLIFRSIIPNRPPTQDTVQALVDDVILPSLT
MMAR_1695|M.marinum_M RGEITQAAITDELWDLLPGYLIFRSIIPNRPPTPHTVQLLVDDFIMPGLT
MUL_1934|M.ulcerans_Agy99 RGEITQAAITDELWDLLPGYLIFRSIIPNRPPTPHTVQLLVDDFIMPGLT
MAV_3866|M.avium_104 RGEISADAISDELWDLLPGYLIFRSIVPTRPPSRRTVQALVDGFLIPGLT
Mflv_4883|M.gilvum_PYR-GCK RGELRDDVRHIVALQLLTSPLHTFALYSEDPIPPEYCR-LIADLVTRAIR
Mvan_1545|M.vanbaalenii_PYR-1 RGELRDDVRPSIALQLLTCPLHTFALYTDRSVDPGYCR-EIAELVTRAVQ
MAB_0300c|M.abscessus_ATCC_199 RGEVRPGVDARTVVLTLGGYVHLWAAHLGQTAD-DLALDELVDLIIDGVS
MSMEG_3410|M.smegmatis_MC2_155 RGELRADLDFELAQDLLAATVIYRFLFAHADLDPALSEHIVDYVLRGIAA
TH_3283|M.thermoresistible__bu AGQAAPDCDPDAVFELIRGATFLRALTHSDEDAEAFCTRITEAVLRAVRS
*: : .:
Mb1581|M.bovis_AF2122/97 RSTG--
Rv1556|M.tuberculosis_H37Rv RSTG--
MMAR_1695|M.marinum_M QATG--
MUL_1934|M.ulcerans_Agy99 QATG--
MAV_3866|M.avium_104 AR----
Mflv_4883|M.gilvum_PYR-GCK AD----
Mvan_1545|M.vanbaalenii_PYR-1 R-----
MAB_0300c|M.abscessus_ATCC_199 AGR---
MSMEG_3410|M.smegmatis_MC2_155 GPTESS
TH_3283|M.thermoresistible__bu ------