For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGLSECLSGVTAPGLALAAGLLLAPHAFAQPALTPAPGPAPPPIAAPAPEPLAAPVAGCPDVEIVFARGT TEAPGLGAMGQAFVEDLRSRLGGKTVGVYAVDYPASPDFPTAVEGIADASTHVQQIAMNCPDTKLVLGGY SQGAAVMGFVTTELIPDGVSLSEVPAPMPPDIADHVAAVALLGAPSDRFMDMIKQPPVTIGALYQPKTLE LCVPGDFVCSPGNDLGAHARYISDGLVVQAADFAVGKLAETPAESGGAPA
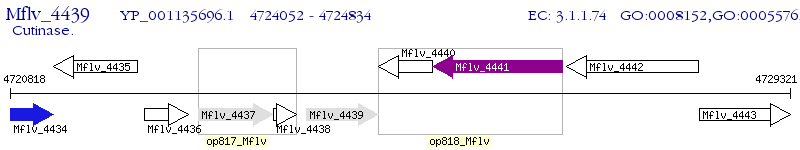
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4439 | - | - | 100% (260) | cutinase |
| M. gilvum PYR-GCK | Mflv_1037 | - | 1e-55 | 49.04% (208) | cutinase |
| M. gilvum PYR-GCK | Mflv_3177 | - | 2e-49 | 44.00% (225) | cutinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1788 | cut1 | 3e-68 | 58.85% (209) | cutinase Cut1 |
| M. tuberculosis H37Rv | Rv1758 | cut1 | 5e-56 | 59.88% (172) | cutinase Cut1 |
| M. leprae Br4923 | MLBr_00099 | - | 2e-10 | 26.25% (259) | hypothetical protein MLBr_00099 |
| M. abscessus ATCC 19977 | MAB_3272c | - | 7e-51 | 51.60% (188) | cutinase Cut1 |
| M. marinum M | MMAR_5251 | cut5 | 7e-62 | 57.65% (196) | cutinase, Cut5 |
| M. avium 104 | MAV_2961 | - | 6e-69 | 60.39% (207) | cutinase Cut3 |
| M. smegmatis MC2 155 | MSMEG_1184 | - | 3e-67 | 61.27% (204) | serine esterase, cutinase family protein |
| M. thermoresistible (build 8) | TH_0238 | - | 4e-55 | 52.79% (197) | serine esterase, cutinase family protein |
| M. ulcerans Agy99 | MUL_4327 | cut5 | 1e-60 | 56.12% (196) | cutinase, Cut5 |
| M. vanbaalenii PYR-1 | Mvan_5804 | - | 2e-59 | 53.98% (226) | cutinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4439|M.gilvum_PYR-GCK ------MGLSECLSGVTAPGLALAAGLLLAPHAFAQPALTPAPGPAPP--
MSMEG_1184|M.smegmatis_MC2_155 ----------MRLAGLVAVAAAATSGMQIA----AMPSAMAAP-------
Mb1788|M.bovis_AF2122/97 ----------------MVTTWALLFAPVPAAS--ADPPDPTVSDGA----
Rv1758|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2961|M.avium_104 ---------------MLSPFWALLLGPALVGT--AAP----AAADS----
MAB_3272c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5804|M.vanbaalenii_PYR-1 -------------MNIFSRWLAVAASVVAVTG--GLPLAAAPAYAAP---
MMAR_5251|M.marinum_M ---------MRNATWTRRLAVG--AATAVTALVGWGIRVPFAAAEP----
MUL_4327|M.ulcerans_Agy99 ---------MRNATWTRRLAVG--AATAVTALVGGGIRVPFVAAEP----
TH_0238|M.thermoresistible__bu ------LSRIGSAVGAALTTASLITGTVITSTVVAGTAVPAAAAQP----
MLBr_00099|M.leprae_Br4923 MVEKVPRKRHRVLAWTAALSMAAVVALAIVAVVILLRSAESPRSSLPPGV
Mflv_4439|M.gilvum_PYR-GCK -----PIAAPAPEPLAAPVAGCPDVEIVFARGTTEAPGLGAMGQ--AFVE
MSMEG_1184|M.smegmatis_MC2_155 ---------------------CPDAEVIFARGTTELPGLGPTGD--AFVE
Mb1788|M.bovis_AF2122/97 ---------------------CPDVEVVFARGTGEPPGVGGIGE--DFID
Rv1758|M.tuberculosis_H37Rv ----------------------------------MP---GRFRE--DFID
MAV_2961|M.avium_104 ---------------------CPDVEVVFARGTSEPPGVGRIGQ--SFVD
MAB_3272c|M.abscessus_ATCC_199 ---------------------------MFARGTFEPPGVGGTGE--SFVN
Mvan_5804|M.vanbaalenii_PYR-1 ---------------------CPDVEVVFARGTFEPPGIGATGQ--AFVD
MMAR_5251|M.marinum_M ---------------------CPDVEVLFARGTGEPPGIGGIGG--SFVA
MUL_4327|M.ulcerans_Agy99 ---------------------CPDVEVLFARGTGEPPGIGGIGG--SFVA
TH_0238|M.thermoresistible__bu ---------------------CPDVQVVFARGTGEPPGVGPTGQ--AFID
MLBr_00099|M.leprae_Br4923 LPIPSTAPHPRKPRPAFQDVSCPDVQLLVVPGTWESSLQDNPLDPVQFPD
. *
Mflv_4439|M.gilvum_PYR-GCK DLR--------SRLGGKTVGVYAVDYPASPDFPTA-----------VEGI
MSMEG_1184|M.smegmatis_MC2_155 ALR--------PQLGDKTLGVYAVDYPATTAFSTA-----------VQGI
Mb1788|M.bovis_AF2122/97 ALR--------SKIGEKSMGVYGVDYPATTDFPTA-----------MAGI
Rv1758|M.tuberculosis_H37Rv ALR--------SKIGEKSMGVYGVDYPATTDFPTA-----------MAGI
MAV_2961|M.avium_104 TLR--------SRLGTKSVGVYPVNYPATTDFPTA-----------ADGI
MAB_3272c|M.abscessus_ATCC_199 ALR--------ARTKGKSVEVYPVEYPASLAFATA-----------ADGV
Mvan_5804|M.vanbaalenii_PYR-1 SLR--------SRAGGRTVDVYAVNYPASFDFATA-----------ADGV
MMAR_5251|M.marinum_M ALR--------SDIGAKSLAVYAVDYPASSDFSSSD-----FPRTVIEGI
MUL_4327|M.ulcerans_Agy99 ALR--------SDIGAKSSAVYAIDYPASSDFSSSD-----FPRTVIDGI
TH_0238|M.thermoresistible__bu ALR--------PRVGDRSLDVYAVNYPAIDVWNTG-----------IDGI
MLBr_00099|M.leprae_Br4923 ALLRNSTMTIGQQFPTSRVQTYTIPYTAQFHNPLSGDKQMTYNDSRAEGT
* .* : *.* . *
Mflv_4439|M.gilvum_PYR-GCK ADASTHVQQIAMNCPDTKLVLGGYSQGAAVMGFVTTEL------------
MSMEG_1184|M.smegmatis_MC2_155 ADARARILANAANCPDTKMVLGGFSQGAAVMGFVTSSA------------
Mb1788|M.bovis_AF2122/97 YDAGTHVEQTAANCPQSKLVLGGFSQGAAVMGFVTAAA------------
Rv1758|M.tuberculosis_H37Rv YDAGTHVEQTAANCPQSKLVLGGFSQGAAVMGFVTAAA------------
MAV_2961|M.avium_104 SDAGAHVQQMAANCPRTKMVLGGYSQGAAVMGFVTESA------------
MAB_3272c|M.abscessus_ATCC_199 IDASNRVKDVAARCPNTKIVMGGFSQGAAVVAYTTADA------------
Mvan_5804|M.vanbaalenii_PYR-1 IDASNRVVTMAANCPDTQLVLGGFSQGAAVAAYITEDT------------
MMAR_5251|M.marinum_M RDASSHIESMAAKCPATREVLGGYSQGAAVAGYVTSAS------------
MUL_4327|M.ulcerans_Agy99 RDASSHIESMAAKCPATREVLGGYSQGAAVAGYVTSAS------------
TH_0238|M.thermoresistible__bu RDAGAHVVRMAEQCPDTRLVLGGFSQGAAVMGFVTSAE------------
MLBr_00099|M.leprae_Br4923 RAMVQEMINVNNKCPLTSYVLVGFSQGAVIAGDITSDIGNGHGPVDDDLV
.: .** : *: *:****.: . *
Mflv_4439|M.gilvum_PYR-GCK -----IPDGVS-LSEVPAPMPPDIADHVAAVALLGAPSDRFMDMIK--QP
MSMEG_1184|M.smegmatis_MC2_155 -----VPDGVS-PTEVPAPMPPEIADHVAAVALFGKPSPRFMRVLN--DP
Mb1788|M.bovis_AF2122/97 -----IPDGAP-LDAP-RPMPPEVADHVAAVTLFGMPSVAFMHSIG--AP
Rv1758|M.tuberculosis_H37Rv -----IPDGAP-LDAP-RPMPPEVADHVAAVTLFGMPSVAFMHSIG--AP
MAV_2961|M.avium_104 -----VPDGVQ-LVQSLQPMPAAVANHVAAVALFGKPSAQFMSIIN--EP
MAB_3272c|M.abscessus_ATCC_199 -----VPPGFELPEGITGPMPAEVADHVAAVALFGKPSSGFLQRIDNTAP
Mvan_5804|M.vanbaalenii_PYR-1 -----IPEGYTPPPGMTGPMPPSVADNVAAVALFGKPSSGFLQMIYTGAP
MMAR_5251|M.marinum_M -----VPPGVP-AAAVPKPMAPDIADHVAAVTLFGTPSEQFLQHYG--AP
MUL_4327|M.ulcerans_Agy99 -----VPPGVP-AAAVPKPMAPDIADYVAAVTLFGTPSEQFLQHYG--AP
TH_0238|M.thermoresistible__bu -----VPAGVD-PATVPKPLDPDIAEHVAAVVLFGMPNARAMNFLG--TP
MLBr_00099|M.leprae_Br4923 LGVTLIADGRR-QQGVGNDIGPNPPGEGAEVTLHEVPVLSGLGMTMTGAR
:. * : . . * *.* * :
Mflv_4439|M.gilvum_PYR-GCK PVTIGALYQPKTLELCVPGDFVCSP-GNDLGAHARYISDG-LVVQA----
MSMEG_1184|M.smegmatis_MC2_155 PVVVGPYYASKAIDLCVENDLVCDPKGSSFAVHNQYQEFG-LVTQA----
Mb1788|M.bovis_AF2122/97 PIVIGPLYAEKTIQLCAPGDPVCSS-GGNWAAHNGYADDG-MVEQA----
Rv1758|M.tuberculosis_H37Rv PIVIGPLYAEKTIQLCAPGDPVCSS-GGNWAAHNGYADDG-MVEQA----
MAV_2961|M.avium_104 PVTIGPLYAAKTIELCVPGDPICSG-AVNPAAHRQYVEAG-MVDQA----
MAB_3272c|M.abscessus_ATCC_199 PINIGHLYVAKTMDMCVPEDPICSPDGSDNAAHGSYGDNG-MTSQA----
Mvan_5804|M.vanbaalenii_PYR-1 PINVGSRYAAKTLDLCIPEDPVCSPTGRDNAAHGGYALTG-MTDRA----
MMAR_5251|M.marinum_M PITIGPLYQPRTVELCASGDPVCSG-GNDAAAHALYAVNG-MTGQG----
MUL_4327|M.ulcerans_Agy99 PITIGPLYQPRTIELCASGDPVCSG-GNDAAAHALYAVNG-MTGQG----
TH_0238|M.thermoresistible__bu PVVIGPLYEDKTVKLCAVDDPVCSN-GMNFAAHNTYATNAAMVDQG----
MLBr_00099|M.leprae_Br4923 PGGFG-VLHSRTNEICAPGDLICAAPAEAFSVANLPATLNTLASGAGQPI
* .* :: .:* * :* . .. :. .
Mflv_4439|M.gilvum_PYR-GCK -ADFAVGKL-----------------------------------------
MSMEG_1184|M.smegmatis_MC2_155 -ATFVAQKLQADWAAEEATDEESTDEATPAPRTPPTRTPSPLSAPVGATP
Mb1788|M.bovis_AF2122/97 -AVFAAGRLG----------------------------------------
Rv1758|M.tuberculosis_H37Rv -AVFAAGRLG----------------------------------------
MAV_2961|M.avium_104 -VDFTTDRV-----------------------------------------
MAB_3272c|M.abscessus_ATCC_199 -ADFAARQIN----------------------------------------
Mvan_5804|M.vanbaalenii_PYR-1 -AEFAVRNMPRHG-------------------------------------
MMAR_5251|M.marinum_M -ADFAAHHL-----------------------------------------
MUL_4327|M.ulcerans_Agy99 -ADFAAHHL-----------------------------------------
TH_0238|M.thermoresistible__bu -AAFAADRLG----------------------------------------
MLBr_00099|M.leprae_Br4923 HAMYATAQFWDLDG-------------------------------APATV
. :.. ..
Mflv_4439|M.gilvum_PYR-GCK -----AETPAESGGAPA--------------
MSMEG_1184|M.smegmatis_MC2_155 PVQQIASTPAPVGTGPLPGPAVPPPPTAPLA
Mb1788|M.bovis_AF2122/97 -------------------------------
Rv1758|M.tuberculosis_H37Rv -------------------------------
MAV_2961|M.avium_104 -------------------------------
MAB_3272c|M.abscessus_ATCC_199 -------SPTDTVSAAG--------------
Mvan_5804|M.vanbaalenii_PYR-1 -----SEQPVDLAAGPR--------------
MMAR_5251|M.marinum_M -------------------------------
MUL_4327|M.ulcerans_Agy99 -------------------------------
TH_0238|M.thermoresistible__bu ---VASDKPLPTPSSGQNFGN----------
MLBr_00099|M.leprae_Br4923 WTLNWVHRLIEGAPHPKHG------------