For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRNATWTRRLAVGAATAVTALVGWGIRVPFAAAEPCPDVEVLFARGTGEPPGIGGIGGSFVAALRSDIGA KSLAVYAVDYPASSDFSSSDFPRTVIEGIRDASSHIESMAAKCPATREVLGGYSQGAAVAGYVTSASVPP GVPAAAVPKPMAPDIADHVAAVTLFGTPSEQFLQHYGAPPITIGPLYQPRTVELCASGDPVCSGGNDAAA HALYAVNGMTGQGADFAAHHL
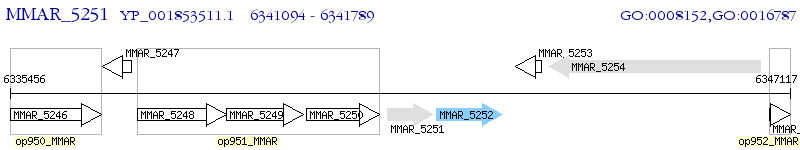
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5251 | cut5 | - | 100% (231) | cutinase, Cut5 |
| M. marinum M | MMAR_2419 | cut1 | 1e-63 | 54.50% (222) | cutinase Cut1 |
| M. marinum M | MMAR_3837 | - | 1e-51 | 45.73% (234) | cutinase precursor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3751 | cut5 | 2e-93 | 71.93% (228) | cutinase precursor CUT5 |
| M. gilvum PYR-GCK | Mflv_1037 | - | 2e-67 | 54.79% (219) | cutinase |
| M. tuberculosis H37Rv | Rv3724B | cut5b | 8e-71 | 74.10% (166) | cutinase |
| M. leprae Br4923 | MLBr_00099 | - | 7e-08 | 27.05% (207) | hypothetical protein MLBr_00099 |
| M. abscessus ATCC 19977 | MAB_3272c | - | 5e-49 | 51.55% (194) | cutinase Cut1 |
| M. avium 104 | MAV_0369 | - | 2e-87 | 70.09% (224) | serine esterase cutinase |
| M. smegmatis MC2 155 | MSMEG_5878 | - | 2e-61 | 53.36% (223) | serine esterase, cutinase family protein |
| M. thermoresistible (build 8) | TH_1243 | - | 4e-67 | 57.94% (214) | PROBABLE CUTINASE |
| M. ulcerans Agy99 | MUL_4327 | cut5 | 1e-129 | 96.97% (231) | cutinase, Cut5 |
| M. vanbaalenii PYR-1 | Mvan_5793 | - | 2e-71 | 57.08% (226) | cutinase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5251|M.marinum_M --------------MRNATWTRRLAVGAATAVTA---------LVGWG--
MUL_4327|M.ulcerans_Agy99 --------------MRNATWTRRLAVGAATAVTA---------LVGGG--
Mb3751|M.bovis_AF2122/97 --------------MDVIRWARRLAVVAGTAAAV---------TTPGLLS
Rv3724B|M.tuberculosis_H37Rv -------------------------------------------MAPG---
MAV_0369|M.avium_104 -----------------MSFQVRAALTVLTAAAA---------LCVAP-M
Mflv_1037|M.gilvum_PYR-GCK -----------------MEMVNRIAGIAGATVLG-------AVSAVVGVG
Mvan_5793|M.vanbaalenii_PYR-1 -----------------MKMS-RVAGIAGAVALG-------ACSAVTGLG
MSMEG_5878|M.smegmatis_MC2_155 -----------------MNVLKLLG--AALVAFG-------ALPVIPAS-
TH_1243|M.thermoresistible__bu --MGWVDMRVTELPNRSRSMFRLIGVISAFVLAGSMACGIGFVSAVMGVG
MAB_3272c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00099|M.leprae_Br4923 MVEKVPRKRHRVLAWTAALSMAAVVALAIVAVVILLRSAESPRSSLPPGV
MMAR_5251|M.marinum_M IRVPFAAAEP-----------CPDVEVLFARGTGEPPGIG----------
MUL_4327|M.ulcerans_Agy99 IRVPFVAAEP-----------CPDVEVLFARGTGEPPGIG----------
Mb3751|M.bovis_AF2122/97 AHVPMVSAEP-----------CPDVEVVFARGTGEPPGIG----------
Rv3724B|M.tuberculosis_H37Rv SHLVLAASED-----------CSSTHCV----------------------
MAV_0369|M.avium_104 PGAPAAGADP-----------CPDVEVVFARGSGEPPGIG----------
Mflv_1037|M.gilvum_PYR-GCK VTPAASAQQP-----------CPDVEVVFARGTSEAPGVG----------
Mvan_5793|M.vanbaalenii_PYR-1 AP--SAAAQP-----------CPDVEVVFARGTSEPPGVG----------
MSMEG_5878|M.smegmatis_MC2_155 ----PASAQP-----------CPDVEVVFARGTFEPPGVG----------
TH_1243|M.thermoresistible__bu SP--TAAAQP-----------CPDVEVIFARGTDEPPGVG----------
MAB_3272c|M.abscessus_ATCC_199 ---------------------------MFARGTFEPPGVG----------
MLBr_00099|M.leprae_Br4923 LPIPSTAPHPRKPRPAFQDVSCPDVQLLVVPGTWESSLQDNPLDPVQFPD
:
MMAR_5251|M.marinum_M GIGGSFVAALRSDIGAKSLAVYAVDYPASSDFSS-----SDFPRTVIEGI
MUL_4327|M.ulcerans_Agy99 GIGGSFVAALRSDIGAKSSAVYAIDYPASSDFSS-----SDFPRTVIDGI
Mb3751|M.bovis_AF2122/97 SVGGLFVDALRSQVGAKSLGVYAVNYPASNDFAS-----SDFPKTVIDGI
Rv3724B|M.tuberculosis_H37Rv -----------SQVGAKSLGVYAVNYPASNDFAS-----SDFPKTVIDGI
MAV_0369|M.avium_104 GIGQPFVDALRSQIGSRSLAVYAVKYPASTDFSN-----PDFPATVIDGI
Mflv_1037|M.gilvum_PYR-GCK GVGQAFVDAVRAQAAPRTVGVYAVNYPAGSNFGDR----ELFAGTVIDGV
Mvan_5793|M.vanbaalenii_PYR-1 GVGQAFVDALRAQAAPRTVGVYAVNYPAANNFTDR----AAFAGTVIDGI
MSMEG_5878|M.smegmatis_MC2_155 VMGQAFVDTLRAQAAPRSVDVYPVDYPASGDFGDR----IAFARTVVDGI
TH_1243|M.thermoresistible__bu GVGQAFIDELRPRIGDRSLSVYAVNYPASSNFGAG----IEFARTVVDGI
MAB_3272c|M.abscessus_ATCC_199 GTGESFVNALRARTKGKSVEVYPVEYPAS----------LAFA-TAADGV
MLBr_00099|M.leprae_Br4923 ALLRNSTMTIGQQFPTSRVQTYTIPYTAQFHNPLSGDKQMTYNDSRAEGT
.*.: *.* : : :*
MMAR_5251|M.marinum_M RDASSHIESMAAKCPATREVLGGYSQGAAVAGYVTSASVPPGVPAAA---
MUL_4327|M.ulcerans_Agy99 RDASSHIESMAAKCPATREVLGGYSQGAAVAGYVTSASVPPGVPAAA---
Mb3751|M.bovis_AF2122/97 RDAGSHIQSMAMSCPQTRQVLGGYSQGAAVAGYVTSAVVPPAVPVQA---
Rv3724B|M.tuberculosis_H37Rv RDAGSHIQSMAMSCPQTRQVLGGYSQGAAVAGYVTSAVVPPAVPVQA---
MAV_0369|M.avium_104 RDASSHLESMVASCPNTREVLGGYSQGAAVAGYTTSAVVPPGVPASA---
Mflv_1037|M.gilvum_PYR-GCK RDAGNRVLAVAEECPDTRIVLGGFSQGAVVSGFTTSDTVPSSVPAAA---
Mvan_5793|M.vanbaalenii_PYR-1 RDASNRLQAMSVNCPNTRLVLGGFSQGAVVSGFTTSDTVPSSVPAAA---
MSMEG_5878|M.smegmatis_MC2_155 ADAASHVEATADTCPDTRIVVGGYSQGAVVAGFVTAADIPAEIPAEYRQY
TH_1243|M.thermoresistible__bu RDAADRVQTTAANCPDTRIVLGGYSQGAALAGYVTAAAVPEEVPVEFRSY
MAB_3272c|M.abscessus_ATCC_199 IDASNRVKDVAARCPNTKIVMGGFSQGAAVVAYTTADAVPPGFELPEG--
MLBr_00099|M.leprae_Br4923 RAMVQEMINVNNKCPLTSYVLVGFSQGAVIAGDITSDIGNGHGPVDDDLV
..: ** * *: *:****.: . *:
MMAR_5251|M.marinum_M --------------VPKPMAPDIADHVAAVTLFGTPSEQFLQHYG--APP
MUL_4327|M.ulcerans_Agy99 --------------VPKPMAPDIADYVAAVTLFGTPSEQFLQHYG--APP
Mb3751|M.bovis_AF2122/97 --------------VPAPMAPEVANHVAAVTLFGAPSAQFLGQYG--APP
Rv3724B|M.tuberculosis_H37Rv --------------VPAPMAPEVANHVAAVTLFGAPSAQFLGQYG--APP
MAV_0369|M.avium_104 --------------VPPPMPPEVAKHVAAVTLFGTPSGQFLQKYH--APP
Mflv_1037|M.gilvum_PYR-GCK --------------APTPLPPEVADHVAAVVLFGTPSEAFMRRFG--APE
Mvan_5793|M.vanbaalenii_PYR-1 --------------APTPLPPKVAEHVAAVVLFGAPSGSFMEKYG--APA
MSMEG_5878|M.smegmatis_MC2_155 --------------IPDPMPAEVSDHVAAVVLIGKPSDQFMRDIG--APP
TH_1243|M.thermoresistible__bu --------------VPEPMPREVADHVAAVVLFAQPSSALLQRYG--APP
MAB_3272c|M.abscessus_ATCC_199 --------------ITGPMPAEVADHVAAVALFGKPSSGFLQRIDNTAPP
MLBr_00099|M.leprae_Br4923 LGVTLIADGRRQQGVGNDIGPNPPGEGAEVTLHEVPVLSGLGMTMTGARP
: . . * *.* * : *
MMAR_5251|M.marinum_M ITIGPLYQPRTVELCASGDPVCS-------------------GGNDAAAH
MUL_4327|M.ulcerans_Agy99 ITIGPLYQPRTIELCASGDPVCS-------------------GGNDAAAH
Mb3751|M.bovis_AF2122/97 IAIGPLYQPKTLQLCADGDSICG-------------------DGNSPVAH
Rv3724B|M.tuberculosis_H37Rv IAIGPLYQPKTLQLCADGDSICG-------------------DGNSPVAH
MAV_0369|M.avium_104 LAIGPLYQPKTLQLCAVGDPICGT-----------------GGGNDLAAH
Mflv_1037|M.gilvum_PYR-GCK ITIGPLYADKTLKLCAQGDTICDG----------------APDAGPNVAH
Mvan_5793|M.vanbaalenii_PYR-1 VTVGPLYADRTVELCAAGDSICDG----------------APNGGPNLAH
MSMEG_5878|M.smegmatis_MC2_155 IVIGPRYVDKTLNLCAPGDTICDG----------------APAGGPSFAH
TH_1243|M.thermoresistible__bu IAIGSVYQDKTLELCTEGDTICNG----------------VADVMPTFAH
MAB_3272c|M.abscessus_ATCC_199 INIGHLYVAKTMDMCVPEDPICS------------------PDGSDNAAH
MLBr_00099|M.leprae_Br4923 GGFG-VLHSRTNEICAPGDLICAAPAEAFSVANLPATLNTLASGAGQPIH
.* :* .:*. * :* *
MMAR_5251|M.marinum_M ALYAVNGMTGQGADFAAHHL-----------------
MUL_4327|M.ulcerans_Agy99 ALYAVNGMTGQGADFAAHHL-----------------
Mb3751|M.bovis_AF2122/97 GLYAVNGMVGQGANFAASRL-----------------
Rv3724B|M.tuberculosis_H37Rv GLYAVNGMVGQGANFAASRL-----------------
MAV_0369|M.avium_104 TSYPVNGMTNQAAQYVFSHL-----------------
Mflv_1037|M.gilvum_PYR-GCK AFYPVNGMINEGATYAVGRL-----------------
Mvan_5793|M.vanbaalenii_PYR-1 ALYPVNGMVGQGASYAAGRL-----------------
MSMEG_5878|M.smegmatis_MC2_155 ALYNVNGMMGQAAQYVIDRL-----------------
TH_1243|M.thermoresistible__bu VTYTWNGMPAEGAAFAADRL-----------------
MAB_3272c|M.abscessus_ATCC_199 GSYGDNGMTSQAADFAARQINSPTDTVSAAG------
MLBr_00099|M.leprae_Br4923 AMYATAQFWDLDGAPATVWTLNWVHRLIEGAPHPKHG
* : . .