For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEFSNVTALVTGASGGIGEEFAVQLARRGANLILVARRAERLEALRRTLVARHPGIVVDVLTADLSGAGS GAEVEGGVRELGRSVDVLINNAGIGLHGKFVDQEPEANTAQIQLNCGTLVDLTARFLPGMVERRHGVVLN VASTAAFQPTPGMAVYGATKAFVLSYTEALWQECRGTGVRILALCPGATETEFFDRTGEKFLTDGRQTAE QVVDTAFAALDKSDPTVVSGLRNALLASGYRIAPRKLLLAVSERMLRSS
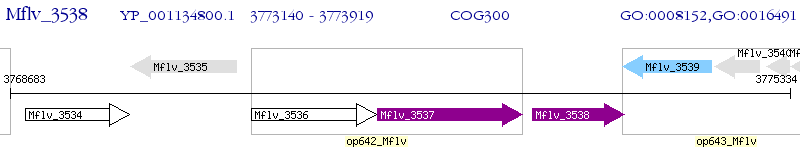
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3538 | - | - | 100% (259) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_0138 | - | 4e-46 | 42.91% (254) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_0244 | - | 2e-33 | 37.15% (253) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1571 | - | 2e-41 | 41.94% (248) | ketoacyl reductase |
| M. tuberculosis H37Rv | Rv1544 | - | 2e-41 | 41.94% (248) | ketoacyl reductase |
| M. leprae Br4923 | MLBr_00429 | - | 7e-31 | 34.54% (249) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_4261c | - | 6e-32 | 37.30% (244) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_0254 | - | 2e-43 | 42.75% (255) | ketoacyl reductase |
| M. avium 104 | MAV_3226 | - | 5e-42 | 41.63% (245) | ketoacyl reductase |
| M. smegmatis MC2 155 | MSMEG_0737 | - | 1e-33 | 37.35% (249) | dehydrogenase |
| M. thermoresistible (build 8) | TH_4326 | - | 7e-37 | 38.65% (251) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1543 | - | 4e-43 | 43.27% (245) | ketoacyl reductase |
| M. vanbaalenii PYR-1 | Mvan_3324 | - | 1e-122 | 83.78% (259) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3538|M.gilvum_PYR-GCK ---------MEFSNVTALVTGASGGIGEEFAVQLARRGANLILVARRAER
Mvan_3324|M.vanbaalenii_PYR-1 MMAPTEEHRVEFSNVTALVTGASGGIGEEFAVQLARRGANLILVARRAEK
MMAR_0254|M.marinum_M MALP-----PPGKDRAAIVTGASSGIGEEFARILSQRGYQVVLVARSADR
Mb1571|M.bovis_AF2122/97 MSLP-----KPNNQTTVVITGASSGIGVELARGLAGRGFPLMLVARRRER
Rv1544|M.tuberculosis_H37Rv MSLP-----KPNNQTTVVITGASSGIGVELARGLAGRGFPLMLVARRRER
MUL_1543|M.ulcerans_Agy99 MTLP-----KPNKQATVVITGASSGIGVELARGLAGRGFPLMLVARRRER
MAV_3226|M.avium_104 MSLP-----KPDIATTVVITGASSGIGAELARGLARRGFPLLLVARRRER
MLBr_00429|M.leprae_Br4923 MPIP-----APSPEARAVVTGASQNIGEALATELAAHGHSLIVIARREDV
MSMEG_0737|M.smegmatis_MC2_155 -----------MPRPVALITGPTSGLGAGFARRYALEGYDLVFVARDQQR
TH_4326|M.thermoresistible__bu -----------MPRPVALITGPTSGLGEGFARRYAADGHDLVLVARDVQR
MAB_4261c|M.abscessus_ATCC_199 -MAA-----TNSARPVALITGPTSGLGGGFARKYASQGYDVVLVARDEQR
.::**.: .:* :* : * ::.:** :
Mflv_3538|M.gilvum_PYR-GCK LEALRRTLVAR-HPGIVVDVLTADLSGAGSGAEVEGGVRELGRSVDVLIN
Mvan_3324|M.vanbaalenii_PYR-1 LAALQHTLTSR-HPGIVVDVLTVDLALPGSAAELEAAVRELGRTVDVLIN
MMAR_0254|M.marinum_M LEALAGRLGSDTHP------LPADLSVRSDRAGLVDRVAALGLVPDILIN
Mb1571|M.bovis_AF2122/97 LDELADQLRQE--HCVGVEVLPLDLADTQARAQLADRLRSDA--IAGLCN
Rv1544|M.tuberculosis_H37Rv LDELADQLRQE--HCVGVEVLPLDLADTQARAQLADRLRSDA--IAGLCN
MUL_1543|M.ulcerans_Agy99 LEELADDLRDK--FSIAVEVLPLDLSDAQARTQLADRLRSDS--IAGLCN
MAV_3226|M.avium_104 LDDLANEVGGQ--YSVGVEVLPLDLGDSKARARLAGRLRSES--IAGLCN
MLBr_00429|M.leprae_Br4923 LVDLAARLADK--YRVAVEVRPADLTDPSERVKLADELTVRP--ISILCA
MSMEG_0737|M.smegmatis_MC2_155 LEQLADELHHE--AGATVETIAADLAVADDRARVAERVRQG---VRVLVN
TH_4326|M.thermoresistible__bu LEQLAAELRQS--AGIDVEVLPADLAVAADRAKVADRLADG---VQVLVN
MAB_4261c|M.abscessus_ATCC_199 LGALADELTCR--FGVYAEALPADLADAADRARVADRLAAG---VSVLVN
* * : . ** . : : *
Mflv_3538|M.gilvum_PYR-GCK NAGIGLHGKFVDQEPEANTAQIQLNCGTLVDLTARFLPGMVERRHGVVLN
Mvan_3324|M.vanbaalenii_PYR-1 NAGVGLHGKFATQEPAPNAAQIQLNCGTLVDLTARFLPPMIERRHGVVLN
MMAR_0254|M.marinum_M NAGLSTLGPVAKSVPEQELNLVEVDVAAVVDLCSRFLPAMVERGRGAVLN
Mb1571|M.bovis_AF2122/97 SAGFGTSGRFWELPFARESEEVVLNALALMELTHAALPGMVKRGAGAVLN
Rv1544|M.tuberculosis_H37Rv SAGFGTSGRFWELPFARESEEVVLNALALMELTHAALPGMVKRGAGAVLN
MUL_1543|M.ulcerans_Agy99 SAGFGTSGRFCELPPERESEQVTLNALALMEFTRAALPGMVERGAGAVLN
MAV_3226|M.avium_104 SAGFGTSGVFHELPMERESEEVTLNALALMELTHATLPGMVSRGAGAVLN
MLBr_00429|M.leprae_Br4923 NAGTATFGPVSALDPAGEKAQVQLNAVAVHDLTLAVLPGMIERKAGGILI
MSMEG_0737|M.smegmatis_MC2_155 NAGFATSGEFWTADFARLRAQLDVNVTAVMELTHAALPAMLDAHEGTIIN
TH_4326|M.thermoresistible__bu NAGFGTSGEFWTADPTLLQAQLDVNVTAVMHLTHAALPPMLDAHRGTIIN
MAB_4261c|M.abscessus_ATCC_199 NAGFATAGEFWTAAPELLHSQLDVNVTAVMELTRAALPSMIEAGAGTVIN
.** . * . : :: :: .: ** *:. * ::
Mflv_3538|M.gilvum_PYR-GCK VASTAAFQPTPGMAVYGATKAFVLSYTEALWQECRGTGVRILALCPGATE
Mvan_3324|M.vanbaalenii_PYR-1 LASTAAFQPTPGMAVYGATKAFVLSFTEALWQECRGSGVRVLALCPGATE
MMAR_0254|M.marinum_M VASVAGFAPLPGQAAYGAAKAFVLSYTHSLRGELHGSGVSVTALSPGPVD
Mb1571|M.bovis_AF2122/97 IASIAGFQPIPYMAVYSATKAFVLTFSEAVQEELHGTGVSVTALCPGPVP
Rv1544|M.tuberculosis_H37Rv IASIAGFQPIPYMAVYSATKAFVLTFSEAVQEELHGTGVSVTALCPGPVP
MUL_1543|M.ulcerans_Agy99 IASIAGFQPIPFMAVYSASKAFVLTFSEAVQEELHGTGVSVTALCPGPVP
MAV_3226|M.avium_104 IASIAGFQPVPYMAVYSATKAFVQTFSEAVHEELHGSGVSVTCLCPGPVP
MLBr_00429|M.leprae_Br4923 SGSAAGNSPIPYNATYAATKAFANTFSESLRGELHGSGVHVTLLAPGPVR
MSMEG_0737|M.smegmatis_MC2_155 VASVAGLVPG-RGSTYSASKAWVVSFTEGLANGLIGTGVGVHALCPGFVR
TH_4326|M.thermoresistible__bu VASVAGLLPG-RGSTYSASKAWVIAFSEGLANGLTGTGVGIHVVCPGFVR
MAB_4261c|M.abscessus_ATCC_199 VASVAGLVSG-RGSTYSASKAWVVSFTEGLANGLLGTGVGMHVLCPGFIH
.* *. . :.*.*:**:. :::..: *:** : :.**
Mflv_3538|M.gilvum_PYR-GCK TEFFDRTG------EKFLTDGRQTAEQVVDTAFAALDKSDPTVVSGLRNA
Mvan_3324|M.vanbaalenii_PYR-1 TEFFDRTG------EEFLTDGRQTSRQVVDTAFAALDGSSPTVISGFRNK
MMAR_0254|M.marinum_M TSFGDVAGFTQQETESLPKIMWKSVDQVAQAGIDGLAAGKAVVVPGLANR
Mb1571|M.bovis_AF2122/97 TEWAEIASA----ERFSIPLAQVSPHDVAEAAIAGMLSGKRTVVPGIVPK
Rv1544|M.tuberculosis_H37Rv TEWAEIASA----ERFSIPLAQVSPHDVAEAAIAGMLSGKRTVVPGIVPK
MUL_1543|M.ulcerans_Agy99 TEWAEIASA----ERFSIPLARVSPHDVAEAAIEGMLTGKRTVVPGVVPK
MAV_3226|M.avium_104 TEWAEIANA----ERFSIPIAQVSPRDVAEAAIGGMLTGRRTVVPGVVPK
MLBr_00429|M.leprae_Br4923 TELPDHDEASLV-EKLVPDFLWISTEHTARLSLDALERNKMRVVPGLTSK
MSMEG_0737|M.smegmatis_MC2_155 TEFHARAGID---MAGTPSFMWLEVDDVVRDCLADVAKDKVVIVPGLQYK
TH_4326|M.thermoresistible__bu TEFHQRAGID---MSGTPSILYLDVDDVVRTTVARVRKGDVVIVPGLQYK
MAB_4261c|M.abscessus_ATCC_199 TEFHERAGIE---MGTIPEFMWLQVDDVVNACLHDIAAGKVLIVPGVQYK
*. ... . : . ::.*.
Mflv_3538|M.gilvum_PYR-GCK LLASGYRIAPRKLLLAVSERMLRSS------
Mvan_3324|M.vanbaalenii_PYR-1 LLASGYRVSPRKVLLAVSEYMLKANR-----
MMAR_0254|M.marinum_M ISAELIRVTPPDWMGSIMARIHPAMKRQ---
Mb1571|M.bovis_AF2122/97 FVSTSGRFAPRSLLLPAIR-IGNRLRGGPSR
Rv1544|M.tuberculosis_H37Rv FVSTSGRFAPRSLLLPAIR-IGNRLRGGPSR
MUL_1543|M.ulcerans_Agy99 FVSTSGRYLPRSVLLPAIR-IGNKLRGGPSH
MAV_3226|M.avium_104 VVSTGGRFAPRSLLLPGIR-IGNRFRGGPKR
MLBr_00429|M.leprae_Br4923 AMSVASRYAPRAIVASIVGNFYKKLGGG---
MSMEG_0737|M.smegmatis_MC2_155 ALTTGGRMAPRNLVRAVTKVVGKGRART---
TH_4326|M.thermoresistible__bu VLTTVGRLVPRKLVRALTKVVGKGRGRT---
MAB_4261c|M.abscessus_ATCC_199 AITSVTRLIPRGLVRKYNSVVGRGRGRT---
: * * : .