For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSLPKPNNQTTVVITGASSGIGVELARGLAGRGFPLMLVARRRERLDELADQLRQEHCVGVEVLPLDLAD TQARAQLADRLRSDAIAGLCNSAGFGTSGRFWELPFARESEEVVLNALALMELTHAALPGMVKRGAGAVL NIASIAGFQPIPYMAVYSATKAFVLTFSEAVQEELHGTGVSVTALCPGPVPTEWAEIASAERFSIPLAQV SPHDVAEAAIAGMLSGKRTVVPGIVPKFVSTSGRFAPRSLLLPAIRIGNRLRGGPSR
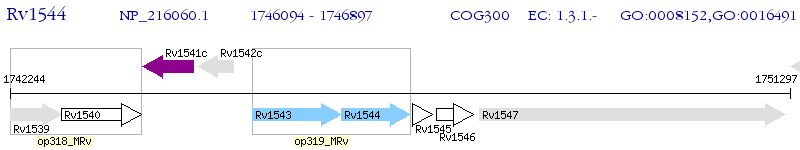
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1544 | - | - | 100% (267) | Possible ketoacyl reductase |
| M. tuberculosis H37Rv | Rv2509 | - | 3e-52 | 43.07% (267) | short-chain type dehydrogenase/reductase |
| M. tuberculosis H37Rv | Rv0765c | - | 6e-25 | 37.32% (209) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1571 | - | 1e-149 | 100.00% (267) | ketoacyl reductase |
| M. gilvum PYR-GCK | Mflv_0138 | - | 1e-49 | 44.11% (263) | short-chain dehydrogenase/reductase SDR |
| M. leprae Br4923 | MLBr_00429 | - | 7e-50 | 42.96% (270) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1537c | - | 1e-47 | 43.41% (258) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_2367 | - | 1e-130 | 88.35% (266) | ketoacyl reductase |
| M. avium 104 | MAV_3226 | - | 1e-123 | 82.77% (267) | ketoacyl reductase |
| M. smegmatis MC2 155 | MSMEG_4722 | - | 2e-46 | 41.79% (268) | short-chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1383 | - | 6e-49 | 42.32% (267) | PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE |
| M. ulcerans Agy99 | MUL_1543 | - | 1e-130 | 87.59% (266) | ketoacyl reductase |
| M. vanbaalenii PYR-1 | Mvan_3113 | - | 5e-85 | 63.81% (257) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1544|M.tuberculosis_H37Rv MSLPKPNNQTTVVITGASSGIGVELARGLAGRGFPLMLVARRRERLDELA
Mb1571|M.bovis_AF2122/97 MSLPKPNNQTTVVITGASSGIGVELARGLAGRGFPLMLVARRRERLDELA
MMAR_2367|M.marinum_M MSLPKPNKQATVVITGASSGIGVELARGLAGRGFPLMLVARRRERLEELA
MUL_1543|M.ulcerans_Agy99 MTLPKPNKQATVVITGASSGIGVELARGLAGRGFPLMLVARRRERLEELA
MAV_3226|M.avium_104 MSLPKPDIATTVVITGASSGIGAELARGLARRGFPLLLVARRRERLDDLA
Mvan_3113|M.vanbaalenii_PYR-1 MSLAKPSAGTTVVITGASSGIGEHIARGLARRGYSLTLIARRRDRLEALS
Mflv_0138|M.gilvum_PYR-GCK MGLPTPSPTTTAVVTGASSGIGADLARELAARGHGVTLVARREDRLRALA
MSMEG_4722|M.smegmatis_MC2_155 MPVPAPSPDARAVVTGASQNIGEALATELAARGHHLIITARREEVLTSLA
TH_1383|M.thermoresistible__bu MPVPEPTPDARAVITGASQNIGAALATELAARGHNLIVTARRAEVLTELA
MLBr_00429|M.leprae_Br4923 MPIPAPSPEARAVVTGASQNIGEALATELAAHGHSLIVIARREDVLVDLA
MAB_1537c|M.abscessus_ATCC_199 MPIPAPSPDARAVVTGASQGIGAALAEELAARGHNLIITARRGEILNDLA
* :. * : .*:****..** :* ** :*. : : *** : * *:
Rv1544|M.tuberculosis_H37Rv DQLRQEHCVGVEVLPLDLADTQARAQLADRLRSD--AIAGLCNSAGFGTS
Mb1571|M.bovis_AF2122/97 DQLRQEHCVGVEVLPLDLADTQARAQLADRLRSD--AIAGLCNSAGFGTS
MMAR_2367|M.marinum_M DDLRDKFSIAVEVLPLDLSDAQARTQLADRLRSD--SIAGLCNSAGFGTS
MUL_1543|M.ulcerans_Agy99 DDLRDKFSIAVEVLPLDLSDAQARTQLADRLRSD--SIAGLCNSAGFGTS
MAV_3226|M.avium_104 NEVGGQYSVGVEVLPLDLGDSKARARLAGRLRSE--SIAGLCNSAGFGTS
Mvan_3113|M.vanbaalenii_PYR-1 DELCRARSISVDVVALDLNEQSARAEAVERLRSG--AVAGLVNSAGFGTN
Mflv_0138|M.gilvum_PYR-GCK DELSSS-NVRVEVVACDVADADARAGLFPEIERRGLTVDVLVNNAGIGTI
MSMEG_4722|M.smegmatis_MC2_155 QRLTERYGVTVEVRAVDLTDPAARATLCDELAER--EISILCANAGTATF
TH_1383|M.thermoresistible__bu DRLTDRHGVTVEVRPLDLADPEQRGTLCAELAER--EISILCANAGVATF
MLBr_00429|M.leprae_Br4923 ARLADKYRVAVEVRPADLTDPSERVKLADELTVR--PISILCANAGTATF
MAB_1537c|M.abscessus_ATCC_199 NALSEKTGVIVEVRAVDLADPGARDALCTELSER--NISILCNNAGTATF
: : *:* . *: : * .: : * .** .*
Rv1544|M.tuberculosis_H37Rv GRFWELPFARESEEVVLNALALMELTHAALPGMVKRGAGAVLNIASIAGF
Mb1571|M.bovis_AF2122/97 GRFWELPFARESEEVVLNALALMELTHAALPGMVKRGAGAVLNIASIAGF
MMAR_2367|M.marinum_M GRFCELPPERESEQVTLNALALMEFTRAALPGMVERGAGAVLNIASIAGF
MUL_1543|M.ulcerans_Agy99 GRFCELPPERESEQVTLNALALMEFTRAALPGMVERGAGAVLNIASIAGF
MAV_3226|M.avium_104 GVFHELPMERESEEVTLNALALMELTHATLPGMVSRGAGAVLNIASIAGF
Mvan_3113|M.vanbaalenii_PYR-1 GLLQDLPPDRERDQVIVNVLALTELTHAVLPQMVERGAGAVLNIGSIAGF
Mflv_0138|M.gilvum_PYR-GCK GPVVDSTPEAEIGQVRVNVEAVIDLTTRAVQQMVPRGRGAILNVGSTAGF
MSMEG_4722|M.smegmatis_MC2_155 GAVKDLDPAGEKAQVQLNVLGVHDLVLAVLPGMVARRAGGILISGSAAGN
TH_1383|M.thermoresistible__bu GPVHKLDPAEEKAQVQLNVLGVHDLVLAVLPGMVQRRAGGILISGSAAGN
MLBr_00429|M.leprae_Br4923 GPVSALDPAGEKAQVQLNAVAVHDLTLAVLPGMIERKAGGILISGSAAGN
MAB_1537c|M.abscessus_ATCC_199 GPIRGLDPAGERKQVQLNAVAVHDLTLAVLPGMLARGAGGILISGSAAGN
* . * :* :*. .: ::. .: *: * *.:* .* **
Rv1544|M.tuberculosis_H37Rv QPIPYMAVYSATKAFVLTFSEAVQEELHGTGVSVTALCPGPVPTEWAEIA
Mb1571|M.bovis_AF2122/97 QPIPYMAVYSATKAFVLTFSEAVQEELHGTGVSVTALCPGPVPTEWAEIA
MMAR_2367|M.marinum_M QPIPFMAVYSASKAFVLTFSEAVQEELHGTGVSVTALCPGPVPTEWAEIA
MUL_1543|M.ulcerans_Agy99 QPIPFMAVYSASKAFVLTFSEAVQEELHGTGVSVTALCPGPVPTEWAEIA
MAV_3226|M.avium_104 QPVPYMAVYSATKAFVQTFSEAVHEELHGSGVSVTCLCPGPVPTEWAEIA
Mvan_3113|M.vanbaalenii_PYR-1 QPLPGAAVYSATKAYVQTFSEAVHEGLHGTGVSCTALCPGPVPTEWWEIA
Mflv_0138|M.gilvum_PYR-GCK QPFPGQSGYAATKAFVRTYTDGLRGELAGTGVTVTALHPGPVRTEFLDTA
MSMEG_4722|M.smegmatis_MC2_155 SPIPNNATYAASKAFANTFSESLRGELTGVGVHVTLLAPGPVRTELPDPS
TH_1383|M.thermoresistible__bu SPIPNNATYAATKAFANTFSESLRGELKDLGVHVTLLAPGPVRSDLPDPS
MLBr_00429|M.leprae_Br4923 SPIPYNATYAATKAFANTFSESLRGELHGSGVHVTLLAPGPVRTELPDHD
MAB_1537c|M.abscessus_ATCC_199 SPIPNNATYAATKAFANTFSESLRGELKGTGVHVTLLAPGPVRTVEPDPA
.*.* : *:*:**:. *:::.:: * . ** * * **** : :
Rv1544|M.tuberculosis_H37Rv SAERFSIPLAQ-----VSPHDVAEAAIAGMLSGKRTVVPGIVPKFVSTSG
Mb1571|M.bovis_AF2122/97 SAERFSIPLAQ-----VSPHDVAEAAIAGMLSGKRTVVPGIVPKFVSTSG
MMAR_2367|M.marinum_M SAERFSIPLAQ-----VSPHDVAEAAIEGMLTGKRTVVPGVVPKFVSTSG
MUL_1543|M.ulcerans_Agy99 SAERFSIPLAR-----VSPHDVAEAAIEGMLTGKRTVVPGVVPKFVSTSG
MAV_3226|M.avium_104 NAERFSIPIAQ-----VSPRDVAEAAIGGMLTGRRTVVPGVVPKVVSTGG
Mvan_3113|M.vanbaalenii_PYR-1 -GEKPPGGKVA-----VSVEEVAEAGIAAMREGKRLVVPGLVPKLTGLGG
Mflv_0138|M.gilvum_PYR-GCK GMDERTFASAFPKFLWIESREVAEKGIDGLAADRGSVIPGLQNEIPARVF
MSMEG_4722|M.smegmatis_MC2_155 EQSLVERLIPD--FLWIDTEYTAKLSLDGLEHNKMRVVPGVTSKAMSVAS
TH_1383|M.thermoresistible__bu EASLVDRLVPD--FLWISTEHTAKLSLDGLAANKMRVVPGLTSKAMSVAS
MLBr_00429|M.leprae_Br4923 EASLVEKLVPD--FLWISTEHTARLSLDALERNKMRVVPGLTSKAMSVAS
MAB_1537c|M.abscessus_ATCC_199 EASIVDKVVPD--FLWINGEYTARVSLNALERNKMRVVPGITSKAMSVAA
. :. . .*. .: .: .: *:**: : .
Rv1544|M.tuberculosis_H37Rv RFAPRSLLLPAIRIGN-RLRGGPSR
Mb1571|M.bovis_AF2122/97 RFAPRSLLLPAIRIGN-RLRGGPSR
MMAR_2367|M.marinum_M RYLPRSVLLPAIRIGN-KLRGGPSH
MUL_1543|M.ulcerans_Agy99 RYLPRSVLLPAIRIGN-KLRGGPSH
MAV_3226|M.avium_104 RFAPRSLLLPGIRIGN-RFRGGPKR
Mvan_3113|M.vanbaalenii_PYR-1 RFTPRALLLPALRMAA-ARRR----
Mflv_0138|M.gilvum_PYR-GCK EFLPRRILLPLIRNQHPALRRSRS-
MSMEG_4722|M.smegmatis_MC2_155 GYAPRAIVTPIVGAVYKKLGGG---
TH_1383|M.thermoresistible__bu GYAPRAIVAPIVGAFYKKLGGG---
MLBr_00429|M.leprae_Br4923 RYAPRAIVASIVGNFYKKLGGG---
MAB_1537c|M.abscessus_ATCC_199 GYAPRAVVAPIVGSFYKKLGD----
: ** :: . :