For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRRGRPRSQATHDAILDAVCDILVTGGYSEVSMDRVATAAGVGKHSLYRRWASKAPLVAEAVMNAYGQQS PLLLPDTGDVEADLREWLLGQARYFGVERNAALSRALAVASADDAGDREALDLQLAGPHRDALLRRLHAA AAGGEIRADTDVEAVTEALIGAMLYRIFNRAKTVDETIKVFNGILDALMRGVRCAENVESRGAHRPGGDD RC*
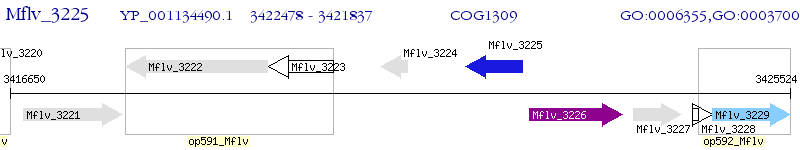
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3225 | - | - | 100% (213) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1535 | - | 5e-19 | 30.57% (193) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4384 | - | 1e-18 | 35.03% (177) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3192c | - | 1e-15 | 35.33% (167) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3167c | - | 1e-15 | 35.33% (167) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00316 | - | 8e-06 | 29.38% (177) | putative TetR/AcrR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3462c | - | 4e-22 | 37.50% (184) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0340 | - | 2e-15 | 32.34% (167) | TetR family transcriptional regulator |
| M. avium 104 | MAV_5165 | - | 2e-16 | 32.95% (173) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_0465 | - | 8e-18 | 36.31% (179) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3283 | - | 5e-17 | 33.85% (195) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_4780 | - | 9e-16 | 32.34% (167) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1961 | - | 8e-18 | 33.33% (177) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3192c|M.bovis_AF2122/97 -----MKADLPSLDKAPGAGRPRDPRIDSAILSATAELLVQIGYSNLSLA
Rv3167c|M.tuberculosis_H37Rv -----MKADLPSLDKAPGAGRPRDPRIDSAILSATAELLVQIGYSNLSLA
MMAR_0340|M.marinum_M --------------MVADFGRPRDPRIDGAVLRATVELLAETGYAGLLVS
MUL_4780|M.ulcerans_Agy99 --------------MVADFGRPRDPRIDGAVLRATVELLAETGYAGLLVS
MAV_5165|M.avium_104 ----------------MDFGRPRDPRIDAAVLRATVELLAETGYPGLLVS
Mvan_1961|M.vanbaalenii_PYR-1 ------------------MGRPRNRAIDDAVLRATVELISESSYAELSLD
TH_3283|M.thermoresistible__bu ----------MPGSGARSPGRPRDPALDDAIRAAARDVVVEHGYDATTLA
Mflv_3225|M.gilvum_PYR-GCK ---------------MNRRGRPRSQATHDAILDAVCDILVTGGYSEVSMD
MSMEG_0465|M.smegmatis_MC2_155 -------MKATDSKLRQRTDGRLDRTRDPAILDAALAALAENGYAGTNMD
MAB_3462c|M.abscessus_ATCC_199 MNETVGPGEVDTPRRHFGNRHGRSEAAREAVLHAADDLLVARGYAGVTME
MLBr_00316|M.leprae_Br4923 -MSVPRSGLSVQMAQPARPLRADAARNRARILGVAYEAFATEGLS-VPID
: .. . . :
Mb3192c|M.bovis_AF2122/97 AVAERAGTTKSALYRRWSSKAELVHEAAFPAA---PTALQAAAGDIAADI
Rv3167c|M.tuberculosis_H37Rv AVAERAGTTKSALYRRWSSKAELVHEAAFPAA---PTALQAAAGDIAADI
MMAR_0340|M.marinum_M AIAKRAGTSKPAIYRRWPSKAHLVHEAVFPID---AATAVPDTGSPVDDL
MUL_4780|M.ulcerans_Agy99 AIAKRAGTSKPAIYRRWPSKAHLVHEAVFPID---AATAVPDTGSPVDDL
MAV_5165|M.avium_104 AIAQRAGTSKPAIYRRWPSKAHLVHEAVFPVG---ADTAIPDTGSTADDL
Mvan_1961|M.vanbaalenii_PYR-1 AIAARAGTSKPAIYRRWRGKAHLVHEAVFPVD---ATIEIPRTGSLHGDV
TH_3283|M.thermoresistible__bu LIADRAGVSVPTIYRRWQHKHELIEEAVLHLD---TFELPEPTGDLEADL
Mflv_3225|M.gilvum_PYR-GCK RVATAAGVGKHSLYRRWASKAPLVAEAVMNAYGQQSPLLLPDTGDVEADL
MSMEG_0465|M.smegmatis_MC2_155 DIASRAGVGKAAIYRRWSSKAALITDALVYWKPALLIDDIPDTGSLEGDL
MAB_3462c|M.abscessus_ATCC_199 GIAKAAGVAKQTVYRWWSSKVDVLMDVFLEDAA--CQLDPPDLGGLEADL
MLBr_00316|M.leprae_Br4923 EIARRAGVGAGTVYRHFPTKEALCAAVIGDRMP--HLVDDGYVLLKSAGP
:* **. ::** : * : . .
Mb3192c|M.bovis_AF2122/97 RMMIAATRDVFTTPVVRAALPGLVADMTADAELNARVLARFADLFAA-VR
Rv3167c|M.tuberculosis_H37Rv RMMIAATRDVFTTPVVRAALPGLVADMTADAELNARVLARFADLFAA-VR
MMAR_0340|M.marinum_M REMVRRTTAVLSTPAAKAALPGLIGEMAADPTLHSALLVRFGEIFGHGLT
MUL_4780|M.ulcerans_Agy99 REMVRRTTAVLSTPAAKAAQPGLIGEMAADPTLHSALLVRFGEIFGHGLT
MAV_5165|M.avium_104 REMVRRTMVFLTTPAAKAALPGLIGEMAADPSLHSALLERFAGAIGGGLA
Mvan_1961|M.vanbaalenii_PYR-1 REMLRRTLAVLSTPAARAALPGLVGEMASDPTLHAALLERFADVLVRDLN
TH_3283|M.thermoresistible__bu GTWVRLFLTVAMEPAARAAVPGLMSAYSRNPDDYRRLMTRGELPVRAAIA
Mflv_3225|M.gilvum_PYR-GCK REWLLGQARYFGVERNAALSRALAVASADDAGDREALDLQLAGPHRDALL
MSMEG_0465|M.smegmatis_MC2_155 NEIAERAARNADERITYDLILRVAVEAMNDSQLASALDDLTLLRGRRTMT
MAB_3462c|M.abscessus_ATCC_199 RHHLSATAQFLAADDAGAVFRALIGQSQHDGQLADDFRGRYLREQQARDQ
MLBr_00316|M.leprae_Br4923 GEALFTYLRSLVLHWGATDRGLVDALAGAGGIDILGVAPDAEDAFLTILS
: . .
Mb3192c|M.bovis_AF2122/97 MRLREAVDRGEAHPDVDPDRLIELIGGATMLRMLLYP---DDMLD---DA
Rv3167c|M.tuberculosis_H37Rv MRLREAVDRGEAHPDVDPDRLIELIGGATMLRMLLYP---DDMLD---DA
MMAR_0340|M.marinum_M TWLDNAAARGQVRADVTAPELAEAIAGIVLIGLLTRP---GELEPGVGDA
MUL_4780|M.ulcerans_Agy99 TWLDNAAARGQVRADVTAPELAEAIAGIVLIGLLTRP---GELEPGVGDA
MAV_5165|M.avium_104 DWLAAAAARGEARPDVTAAELAETIAGVTLVALLTRP---TELD----DA
Mvan_1961|M.vanbaalenii_PYR-1 NYLDSAVRRGEVRRGVTAVELAEAVTGMTLLALITRG---ANVD----DA
TH_3283|M.thermoresistible__bu AMLERAKAAGQAAPDCDPDAVFELIRGATFLRALTHS---DEDA----EA
Mflv_3225|M.gilvum_PYR-GCK RRLHAAAAGGEIRADTDVEAVTEALIGAMLYRIFNRA---KTVDET--IK
MSMEG_0465|M.smegmatis_MC2_155 AILKNATARGEIAPDTDWSLVPDVLTAMLLMRAIRGERVDAN-------Y
MAB_3462c|M.abscessus_ATCC_199 IPIARAVARGELPADVDVAHVAEQLVAPLYYRVIVTG---ETVD----AA
MLBr_00316|M.leprae_Br4923 DLLYAAQYAGTARTDVGVREVKSILVGCQAMEAYNSA-------------
: * * . : . : .
Mb3192c|M.bovis_AF2122/97 WVDQTTAIVVRGVHRAAPGGSVV----------
Rv3167c|M.tuberculosis_H37Rv WVDQTTAIVVRGVHRAAPGGSVV----------
MMAR_0340|M.marinum_M WIDRTTTLLLKGIRKGTPQ--------------
MUL_4780|M.ulcerans_Agy99 WIDRTTTLLLKGIRKGTPQ--------------
MAV_5165|M.avium_104 WVDRTTRLLLKGISA------------------
Mvan_1961|M.vanbaalenii_PYR-1 WIERTAHLITKGISS------------------
TH_3283|M.thermoresistible__bu FCTRITEAVLRAVRS------------------
Mflv_3225|M.gilvum_PYR-GCK VFNGILDALMRGVRCAENVESRGAHRPGGDDRC
MSMEG_0465|M.smegmatis_MC2_155 VRKVIDTVIVPAVRATAY---------------
MAB_3462c|M.abscessus_ATCC_199 FVDRIVEDFLRRRE-------------------
MLBr_00316|M.leprae_Br4923 LAERVTDVVVDGLRAAR----------------
.