For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSVVSVTESYVTLSIVMTTDIGRPRNRAIADAVLRATVELLAESSYAELSLDAVAARAGTSKPAIYRRWR GKAHLVHEAVFPLGADTAIPGSGSLEADVREMTRRTLAVLSTPAALAALPGLVGEMASDPTLHTALLERF GDILVRGLTEFLESAVRRGDVRPDVTASDLAEAVAGMTLLALLTRGTAVDEVWLDRTARLITKGISA*
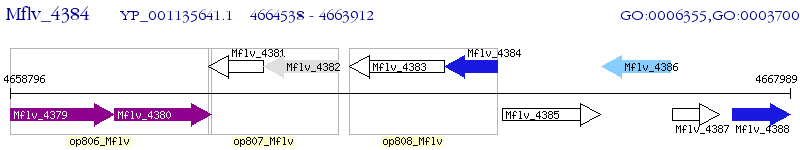
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4384 | - | - | 100% (208) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0492 | - | 1e-39 | 43.75% (192) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4496 | - | 1e-32 | 41.58% (190) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3192c | - | 1e-37 | 44.32% (185) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3167c | - | 1e-37 | 44.32% (185) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3151 | - | 1e-17 | 34.08% (179) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0340 | - | 2e-63 | 64.25% (193) | TetR family transcriptional regulator |
| M. avium 104 | MAV_5165 | - | 4e-67 | 65.96% (188) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2043 | - | 2e-33 | 39.78% (186) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_0688 | - | 1e-32 | 39.18% (194) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_4780 | - | 7e-63 | 63.73% (193) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1961 | - | 2e-79 | 79.14% (187) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3192c|M.bovis_AF2122/97 --------MKADLPSLDKAPGAGRPRDPRIDSAILSATAELLVQIGYSNL
Rv3167c|M.tuberculosis_H37Rv --------MKADLPSLDKAPGAGRPRDPRIDSAILSATAELLVQIGYSNL
MSMEG_2043|M.smegmatis_MC2_155 --------MKAEASPLGKGVGAGRPRDPRIDAAILRATAELLVEIGYANL
TH_0688|M.thermoresistible__bu --------MTTNSTSVDKAAGAGRPRDPRIDGAILRATADLLVEIGYSNL
Mflv_4384|M.gilvum_PYR-GCK MPSVVSVTESYVTLSIVMTTDIGRPRNRAIADAVLRATVELLAESSYAEL
Mvan_1961|M.vanbaalenii_PYR-1 ---------------------MGRPRNRAIDDAVLRATVELISESSYAEL
MMAR_0340|M.marinum_M -----------------MVADFGRPRDPRIDGAVLRATVELLAETGYAGL
MUL_4780|M.ulcerans_Agy99 -----------------MVADFGRPRDPRIDGAVLRATVELLAETGYAGL
MAV_5165|M.avium_104 -------------------MDFGRPRDPRIDAAVLRATVELLAETGYPGL
MAB_3151|M.abscessus_ATCC_1997 ---------MTKNAATQRQRTSGR-LNRSLDIAILEAAFVTLAEQGYDAM
** : : *:* *: : : .* :
Mb3192c|M.bovis_AF2122/97 SLAAVAERAGTTKSALYRRWSSKAELVHEAAFP----AAPTALQAAAGDI
Rv3167c|M.tuberculosis_H37Rv SLAAVAERAGTTKSALYRRWSSKAELVHEAAFP----AAPTALQAAAGDI
MSMEG_2043|M.smegmatis_MC2_155 TMAAVAERAQTTKTALYRRWSSKAELVHEAVFP----AVPTGLVAPAGDI
TH_0688|M.thermoresistible__bu TMSAVAERAGTTKTALYRRWSSKPELVYAATAP----TDPVDTVESSDDV
Mflv_4384|M.gilvum_PYR-GCK SLDAVAARAGTSKPAIYRRWRGKAHLVHEAVFP----LGADTAIPGSGSL
Mvan_1961|M.vanbaalenii_PYR-1 SLDAIAARAGTSKPAIYRRWRGKAHLVHEAVFP----VDATIEIPRTGSL
MMAR_0340|M.marinum_M LVSAIAKRAGTSKPAIYRRWPSKAHLVHEAVFP----IDAATAVPDTGSP
MUL_4780|M.ulcerans_Agy99 LVSAIAKRAGTSKPAIYRRWPSKAHLVHEAVFP----IDAATAVPDTGSP
MAV_5165|M.avium_104 LVSAIAQRAGTSKPAIYRRWPSKAHLVHEAVFP----VGADTAIPDTGST
MAB_3151|M.abscessus_ATCC_1997 SMDNIAARAGVGKAAIYRRWSSKAALTADAILHGRPSLGKLDEVPDTGSL
: :* ** . *.*:**** .*. *. * :..
Mb3192c|M.bovis_AF2122/97 AADIRMMIAATRDVFTTPVVRAALPGLVADMTADAELNARVLARFADLFA
Rv3167c|M.tuberculosis_H37Rv AADIRMMIAATRDVFTTPVVRAALPGLVADMTADAELNARVLARFADLFA
MSMEG_2043|M.smegmatis_MC2_155 AADVHAMIAAARDVFTSPVVRAALPGLIADVSADANLNARVMERFTGVFA
TH_0688|M.thermoresistible__bu AGIVRALIERNRQITATPAARAAMPGLLVDMAADPRLHRGLLGRVGSLVE
Mflv_4384|M.gilvum_PYR-GCK EADVREMTRRTLAVLSTPAALAALPGLVGEMASDPTLHTALLERFGDILV
Mvan_1961|M.vanbaalenii_PYR-1 HGDVREMLRRTLAVLSTPAARAALPGLVGEMASDPTLHAALLERFADVLV
MMAR_0340|M.marinum_M VDDLREMVRRTTAVLSTPAAKAALPGLIGEMAADPTLHSALLVRFGEIFG
MUL_4780|M.ulcerans_Agy99 VDDLREMVRRTTAVLSTPAAKAAQPGLIGEMAADPTLHSALLVRFGEIFG
MAV_5165|M.avium_104 ADDLREMVRRTMVFLTTPAAKAALPGLIGEMAADPSLHSALLERFAGAIG
MAB_3151|M.abscessus_ATCC_1997 RGDLHSLYEARDFGDDQVLTGNLLAGIIAEAVNDPTLAAALDDLLVSQTH
:: : . .*:: : *. * : .
Mb3192c|M.bovis_AF2122/97 A-VRMRLREAVDRGEAHPDVDPDRLIELIGGATMLRMLLYPDDMLD---D
Rv3167c|M.tuberculosis_H37Rv A-VRMRLREAVDRGEAHPDVDPDRLIELIGGATMLRMLLYPDDMLD---D
MSMEG_2043|M.smegmatis_MC2_155 A-VRERIVDGIARGEVHAAVSPDRLVEIIGGATMLRLLLVSDENLD---D
TH_0688|M.thermoresistible__bu A-LRRRLAEAAGRGEVQSGVDADRLTELIGGSVMFRMVLHPDEPIT---E
Mflv_4384|M.gilvum_PYR-GCK RGLTEFLESAVRRGDVRPDVTASDLAEAVAGMTLLALLTRGTAVD----E
Mvan_1961|M.vanbaalenii_PYR-1 RDLNNYLDSAVRRGEVRRGVTAVELAEAVTGMTLLALITRGANVD----D
MMAR_0340|M.marinum_M HGLTTWLDNAAARGQVRADVTAPELAEAIAGIVLIGLLTRPGELEPGVGD
MUL_4780|M.ulcerans_Agy99 HGLTTWLDNAAARGQVRADVTAPELAEAIAGIVLIGLLTRPGELEPGVGD
MAV_5165|M.avium_104 GGLADWLAAAAARGEARPDVTAAELAETIAGVTLVALLTRPTELD----D
MAB_3151|M.abscessus_ATCC_1997 KRMELVFARAVERGEITQGRDLSLVYDIPLGLSLTAALKR-TVIDETYMH
: : . **: : : * : : .
Mb3192c|M.bovis_AF2122/97 AWVDQTTAIVVRGVHRAAPGGSVV
Rv3167c|M.tuberculosis_H37Rv AWVDQTTAIVVRGVHRAAPGGSVV
MSMEG_2043|M.smegmatis_MC2_155 EWVHQTAAIVIHGVTVGR------
TH_0688|M.thermoresistible__bu RWADEVAAILIRGVTKEG------
Mflv_4384|M.gilvum_PYR-GCK VWLDRTARLITKGISA--------
Mvan_1961|M.vanbaalenii_PYR-1 AWIERTAHLITKGISS--------
MMAR_0340|M.marinum_M AWIDRTTTLLLKGIRKGTPQ----
MUL_4780|M.ulcerans_Agy99 AWIDRTTTLLLKGIRKGTPQ----
MAV_5165|M.avium_104 AWVDRTTRLLLKGISA--------
MAB_3151|M.abscessus_ATCC_1997 RMVEEVILPLVQAPTTHT------
... : :.