For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEQSIWMQKVAADPGHSRWYIERFRAMARAGEDLVGEARLVDSIAPRGAHILDAGCGPGRLGRHLAAAGH RVVGVDVDPALIEAAEQDYPGPQWLVGDLAELDLPARGIAEPFDVIVSAGNVMTFLAPSTRARVLGRLRA HLCSDGRAVIGFGAGRDYAFADFLDDAQAAGLVPDLLLSTWDLRPFTDDCDFLVALLRPA*
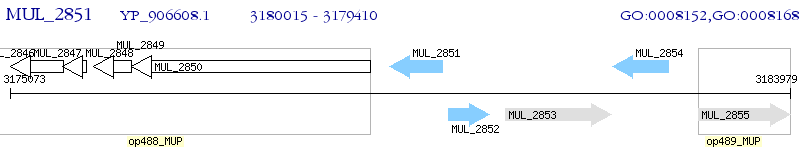
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2851 | - | - | 100% (201) | methyltransferase |
| M. ulcerans Agy99 | MUL_1921 | - | 2e-06 | 31.91% (141) | hypothetical protein MUL_1921 |
| M. ulcerans Agy99 | MUL_0662 | - | 2e-06 | 31.37% (102) | methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2010 | - | 5e-08 | 35.00% (100) | methyltransferase |
| M. gilvum PYR-GCK | Mflv_4890 | - | 2e-92 | 80.90% (199) | methyltransferase type 12 |
| M. tuberculosis H37Rv | Rv1988 | - | 5e-08 | 35.00% (100) | methyltransferase |
| M. leprae Br4923 | MLBr_01713 | - | 1e-05 | 32.20% (118) | hypothetical protein MLBr_01713 |
| M. abscessus ATCC 19977 | MAB_2548c | - | 1e-92 | 81.41% (199) | hypothetical protein MAB_2548c |
| M. marinum M | MMAR_1164 | - | 1e-115 | 100.00% (201) | methyltransferase |
| M. avium 104 | MAV_4332 | - | 3e-96 | 83.08% (201) | SAM-dependent methyltransferases |
| M. smegmatis MC2 155 | MSMEG_3430 | - | 4e-96 | 82.09% (201) | SAM-dependent methyltransferase |
| M. thermoresistible (build 8) | TH_1247 | - | 8e-96 | 83.58% (201) | SAM-dependent methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1534 | - | 3e-99 | 85.57% (201) | methyltransferase type 12 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2851|M.ulcerans_Agy99 ----MVEQSIWMQKVAADPGHSRWYIERFRAMARAGEDLVGEARLVD---
MMAR_1164|M.marinum_M ----MVEQSIWMQKVAADPGHSRWYIERFRAMARAGEDLVGEARLVD---
MSMEG_3430|M.smegmatis_MC2_155 ----MVEQSLWMQKVAADPGHSQWYIERFRSLARAGEDLFGEARLVD---
TH_1247|M.thermoresistible__bu ----MVEKSLWMRKVEADPGHSQWYIERFRAMARAGEDLVGEARLVD---
MAV_4332|M.avium_104 ----MVEQSIWMQKVAADPGHSHWYIERFRAMARAGDDLAGEARFVD---
Mvan_1534|M.vanbaalenii_PYR-1 ----MVETSIWMQKVAADPGHSRWYIERFRAMARAGDDLAGEARLVD---
MAB_2548c|M.abscessus_ATCC_199 ----MVEQSLWMQKVAADPNHSQWYIDRFRSMARDGADLAGEARLVD---
Mflv_4890|M.gilvum_PYR-GCK ----MAETSLWMQKVAADPGHSRWYIERFRAMARAGDDLFGEARLID---
Mb2010|M.bovis_AF2122/97 -----------MSALGRSR--------RAWGWHRLHDEWAARVVSAA---
Rv1988|M.tuberculosis_H37Rv -----------MSALGRSR--------RAWGWHRLHDEWAARVVSAA---
MLBr_01713|M.leprae_Br4923 MSAFVPDVPNRIPDDPSSIDSTAFHGMLTLTGERTIPDLDIENYWFRRHQ
: . * : .
MUL_2851|M.ulcerans_Agy99 ------SIAPRGAHILDAGCGPGRLGRHLAAAGHRVVGVDVDPALIEAAE
MMAR_1164|M.marinum_M ------SIAPRGAHILDAGCGPGRLGRHLAAAGHRVVGVDVDPALIEAAE
MSMEG_3430|M.smegmatis_MC2_155 ------AMAPRNAHILDAGCGPGRLGGYLAGVGHRVVGVDVDPALIAAAE
TH_1247|M.thermoresistible__bu ------AMAPRNAHILDAGCGPGRLGGYLSAAGHRVVGVDVDPALIEAAE
MAV_4332|M.avium_104 ------AMAPRGARILDAGCGPGRLGSYLAAAGHQVVGVDVDPALIEAAE
Mvan_1534|M.vanbaalenii_PYR-1 ------AMAPRGARILDAGCGPGRVGGYLAAAGHQVVGVDVDPALIEAAE
MAB_2548c|M.abscessus_ATCC_199 ------ALVPRGARILDAGCGAGRVGGFLAAAGHHVVGVDVDPALIEAAE
Mflv_4890|M.gilvum_PYR-GCK ------AMVPRGARILDAGCGPGRLGGYLAAAGHHVVGVDVDPVLIDAAE
Mb2010|M.bovis_AF2122/97 ------AVRP-GELVFDIGAGEGALTAHLVRAGARVVAVELHPRRVGVLR
Rv1988|M.tuberculosis_H37Rv ------AVRP-GELVFDIGAGEGALTAHLVRAGARVVAVELHPRRVGVLR
MLBr_01713|M.leprae_Br4923 VVYQWLAQRCTGCEVLEAGCGEGYGADLIVDVAYRVIAVDYDEATVAHVR
: . ::: *.* * : .. :*:.*: . : .
MUL_2851|M.ulcerans_Agy99 QDYPGPQWLVGDLAELDLPARGIAEPFDVIVSAGNVMTFLAPSTRARVLG
MMAR_1164|M.marinum_M QDYPGPQWLVGDLAELDLPARGIAEPFDVIVSAGNVMTFLAPSTRARVLG
MSMEG_3430|M.smegmatis_MC2_155 EDYPGPRWLVGDLAELDLPARGIAEPFDIIVSAGNVMTFLAPSTRAQVLS
TH_1247|M.thermoresistible__bu QDYPGPQYLVGDLAELDLPARGIPDPFDVIVMAGNVMAFLAPSTRTTVLS
MAV_4332|M.avium_104 HDHPGPRWLVGDLAELDLPARGITDPFDVIVSAGNVMTFLAPSTRVLVLS
Mvan_1534|M.vanbaalenii_PYR-1 QDYPGPRWLVGDLAELDLPARGITEPFDLIVSAGNVMTFLAPSTRVQVLA
MAB_2548c|M.abscessus_ATCC_199 HDHPGPRWLVGDLAELDLPSRGISEPFDVIVSAGNVMAFLAPSTRRQVLR
Mflv_4890|M.gilvum_PYR-GCK KDHPGATWLVGDLAELDLPARGIAEPFDVIVSAGNVMTFLAPSTRGEVLR
Mb2010|M.bovis_AF2122/97 ERFPGITVVHADAASIRLPGR----PFRVVANP-------PYGISSRLLR
Rv1988|M.tuberculosis_H37Rv ERFPGITVVHADAASIRLPGR----PFRVVANP-------PYGISSRLLR
MLBr_01713|M.leprae_Br4923 SRYPRVQVMQANLIELPLADASVDVVVNFQVIEHLWNQAQFVSECARVLR
.* : .: .: *. . . . . :*
MUL_2851|M.ulcerans_Agy99 RLRAHLCSDGRAVIGFGAGRDYAFADFLDDAQAAGLVPDLLLST------
MMAR_1164|M.marinum_M RLRAHLCSDGRAVIGFGAGRDYAFADFLDDAQAAGLVPDLLLST------
MSMEG_3430|M.smegmatis_MC2_155 RLRAHLTDNGRAVIGFGAGRDYGFEEFLADAKGAGFTPDQLLST------
TH_1247|M.thermoresistible__bu RLRAHLAEDGRAAIGFGAGRDYDFDEFLADAATAGLTPDLLLST------
MAV_4332|M.avium_104 RLRAHLAADGRAAIGFGAYREYEFTDFLNDAADAGFAPDLLLSS------
Mvan_1534|M.vanbaalenii_PYR-1 RLRAHLRDDGRAVIGFGAGREYEYPQFLDDAAAGGLTPDLLLST------
MAB_2548c|M.abscessus_ATCC_199 RLRSHLADPGRAVIGFGAGRDYEFSEFLSDADAAGLVPDLLLST------
Mflv_4890|M.gilvum_PYR-GCK RLRTHLAAEGRAVIGFGADRDYDFAEFLDDAAGAGFVPDLLLST------
Mb2010|M.bovis_AF2122/97 TLLAPNS--GLVAADLVLQRALVCKFASRNARRFTLTVGLMLPR------
Rv1988|M.tuberculosis_H37Rv TLLAPNS--GLVAADLVLQRALVCKFASRNARRFTLTVGLMLPR------
MLBr_01713|M.leprae_Br4923 -PSGLLMVSTPNRITFSPGRDTSINPFHTRELNADELTQLLITADFSEVA
: * ::.
MUL_2851|M.ulcerans_Agy99 ---------WDLRPFTDDCDFLVALLRPA---------------------
MMAR_1164|M.marinum_M ---------WDLRPFTDDCDFLVALLRPA---------------------
MSMEG_3430|M.smegmatis_MC2_155 ---------WDVRPFSEDSDFLVAILRRA---------------------
TH_1247|M.thermoresistible__bu ---------WDLRPFTEDSEFLVALLRRA---------------------
MAV_4332|M.avium_104 ---------WDLRPFTEDSDFLVAVLRPA---------------------
Mvan_1534|M.vanbaalenii_PYR-1 ---------WDVRPFTDDSDFLVAVLRPA---------------------
MAB_2548c|M.abscessus_ATCC_199 ---------WDLRPFTDKSDFLVALLRRDDAPS-----------------
Mflv_4890|M.gilvum_PYR-GCK ---------WDLRPWREDSGFLVAVLRRM---------------------
Mb2010|M.bovis_AF2122/97 ---------RAFLPPPHVDSAVLVVRRRKCGDWQGR--------------
Rv1988|M.tuberculosis_H37Rv ---------RAFLPPPHVDSAVLVVRRRKCGDWQGR--------------
MLBr_01713|M.leprae_Br4923 MYGLFHGPRLKEMDARHGGSIIDAQIARAVSNAPWPPELVADVAAVTAAD
. : .
MUL_2851|M.ulcerans_Agy99 -------------------------------
MMAR_1164|M.marinum_M -------------------------------
MSMEG_3430|M.smegmatis_MC2_155 -------------------------------
TH_1247|M.thermoresistible__bu -------------------------------
MAV_4332|M.avium_104 -------------------------------
Mvan_1534|M.vanbaalenii_PYR-1 -------------------------------
MAB_2548c|M.abscessus_ATCC_199 -------------------------------
Mflv_4890|M.gilvum_PYR-GCK -------------------------------
Mb2010|M.bovis_AF2122/97 -------------------------------
Rv1988|M.tuberculosis_H37Rv -------------------------------
MLBr_01713|M.leprae_Br4923 FDIIEASAGPEASDGRDVDESLDLIAIAVRR