For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGSLDGRVAFITGVARGQGRSHAIRLASEGAEIIGVDICADIEANRYPLASRAELDETVALVESAGGKML GSVADVRDFGALKSAVDAGVEHFGRLDIVCANAGIAPTAFREVSIEEDLEMWTAAIDVNLVGSYHTAKAA IPHLIAGGQGGAIVFTSSTAGLKGFGGSQGGGLGYAASKHGIVGLMRTLANALAPHSIRVNSVHPTAVNT MMATNPAMTAFLEAYPDGGPHLQNAMPVSLLEPEDISAAISYLVSDAAKYVTGVTFPVDAGFCNKL
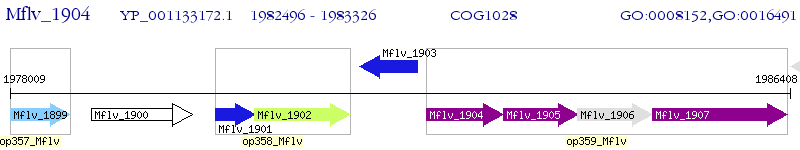
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1904 | - | - | 100% (276) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3996 | - | 1e-78 | 55.91% (279) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1256 | - | 6e-73 | 51.69% (267) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 4e-75 | 54.21% (273) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. tuberculosis H37Rv | Rv2750 | - | 4e-75 | 54.21% (273) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-21 | 30.97% (268) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_3081 | - | 2e-74 | 52.54% (276) | Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_1965 | fabG | 6e-75 | 52.35% (277) | short chain dehydrogenase |
| M. avium 104 | MAV_3550 | - | 1e-125 | 78.99% (276) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5812 | - | 1e-134 | 84.06% (276) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_0236 | - | 1e-80 | 55.15% (272) | PROBABLE DEHYDROGENASE |
| M. ulcerans Agy99 | MUL_3392 | fabG | 6e-74 | 51.62% (277) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_3394 | - | 1e-133 | 82.97% (276) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2771|M.bovis_AF2122/97 ---MIDRPLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICEQIASV
Rv2750|M.tuberculosis_H37Rv ---MIDRPLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICEQIASV
MMAR_1965|M.marinum_M MSRPLDGPLSGRVAFITGAARGQGRAHALRLARDGADVIAVDLCDQIASV
MUL_3392|M.ulcerans_Agy99 MSRPLDGPLSGRVAFITGAARGQGRAHALRLARDGADVIAVDLCDQIASV
MAB_3081|M.abscessus_ATCC_1997 -----MGKLDGKVAFITGAARGQGRAHAIKLASEGASIIAVDICAQIPSV
MSMEG_5812|M.smegmatis_MC2_155 -----MVSLDGKVAFITGAARGQGRSHAVRLASEGADIIGVDICADIASA
Mvan_3394|M.vanbaalenii_PYR-1 -----MGSLDGRVAFITGVARGQGRSHAVRLAADGADIIGVDICSDIASN
Mflv_1904|M.gilvum_PYR-GCK -----MGSLDGRVAFITGVARGQGRSHAIRLASEGAEIIGVDICADIEAN
MAV_3550|M.avium_104 -----MGSLDGKVAFITGAARGQGRSHAVRLAREGANIIGIDICADIAAN
TH_0236|M.thermoresistible__bu -----MGNLEGKVAFITGAARGQGRAHAVRLASEGADIIALDICADIDSM
MLBr_01807|M.leprae_Br4923 -----MTDAAAKVSAAPGRGK-PAFVSRSILVTGGNRGIGLAVARRLVAD
.:*: .* .: . *. * *.: :. : :
Mb2771|M.bovis_AF2122/97 PYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQAGLDEFGR
Rv2750|M.tuberculosis_H37Rv PYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQAGLDEFGR
MMAR_1965|M.marinum_M PYPLGTAEELATTVKLVEDTGARIVASQADVRDREALAAALQAGIDELGQ
MUL_3392|M.ulcerans_Agy99 PYPLGTAEELATTVKLVEDTGARIVASQADVRDREALAAALQAGIDELGQ
MAB_3081|M.abscessus_ATCC_1997 EYPMATTEDLDLTVKEVDALGRAISAHETDVRDREALQAAFDAGVHDLGP
MSMEG_5812|M.smegmatis_MC2_155 GYPMAGREELDETVALVEAHGGKMMSAVADVRDYGALKTALDAGVDHFGR
Mvan_3394|M.vanbaalenii_PYR-1 GYPMATPAELEETVALVEAQGAKMLGSVADVRDFSAVKGALDAGVEHFGR
Mflv_1904|M.gilvum_PYR-GCK RYPLASRAELDETVALVESAGGKMLGSVADVRDFGALKSAVDAGVEHFGR
MAV_3550|M.avium_104 GYPMASRAELDQTVALVEEAGGKMLGTVADVRDFGQVKAALDAGVEQFGR
TH_0236|M.thermoresistible__bu DYPNASPADLEETAKLVEGTGRSIVARQADVRDADAVDEVVREGVERFGR
MLBr_01807|M.leprae_Br4923 GHRVAVT-------HRASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQGP
: . .. : . ** * : .. . *
Mb2771|M.bovis_AF2122/97 LDIVVANAGIAMMQ-------AGDDGWRDVIDVNLTGVFHTVQVAIPTLI
Rv2750|M.tuberculosis_H37Rv LDIVVANAGIAMMQ-------AGDDGWRDVIDVNLTGVFHTVQVAIPTLI
MMAR_1965|M.marinum_M VDIVVANAGIAPMQ-------SGDDGWRDVIDVNLSGAYYTVEVAIPTMI
MUL_3392|M.ulcerans_Agy99 VDIVVANAGIAPMQ-------SGDDGWRDVIDVNLSGAYYTVEVAIPTMI
MAB_3081|M.abscessus_ATCC_1997 VDIVVANAGIATLTP-----ECGDDGWRDVLDVNLTGVYHTIEVARPSMV
MSMEG_5812|M.smegmatis_MC2_155 LDIVCANAGIAAMAFRELTIEEDLQMWADVVDVNLVGSFHTAKAAIPHLI
Mvan_3394|M.vanbaalenii_PYR-1 LDIVCANAGIATMSFRELTDAEDLEMWTDVLDVNLVGTFHTAKAAIPHLI
Mflv_1904|M.gilvum_PYR-GCK LDIVCANAGIAPTAFREVSIEEDLEMWTAAIDVNLVGSYHTAKAAIPHLI
MAV_3550|M.avium_104 LDIVLANAGIAPLAFRQLSIEEELAQWRAVTGVNLDGAYHTAWAAIPHLL
TH_0236|M.thermoresistible__bu LDIVVANAGIVRLSSDEPSQRR--QIFRDIIDVNLVGVWNTVEAAIPHVI
MLBr_01807|M.leprae_Br4923 VEVLVANAGITSDMLIMR---MSEEQFQKVIDVNLTGVFRVAKLASRNMI
:::: *****. : .*** * : . * ::
Mb2771|M.bovis_AF2122/97 EQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYANHLAP
Rv2750|M.tuberculosis_H37Rv EQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYANHLAP
MMAR_1965|M.marinum_M EQGRGGSIVLISSAAGLVGISSADAGAIGYAASKHALVGLMRVYANLLAP
MUL_3392|M.ulcerans_Agy99 EQGRGGSIVLISSAAGLVGISSADAGAIGYVASKHALVGLMRVYANLLAP
MAB_3081|M.abscessus_ATCC_1997 QRGAGGSIVLTSSVAGLTGMVMDNPGGLAYTAAKHGLVGLMRSYANMLAP
MSMEG_5812|M.smegmatis_MC2_155 AGERGGSIVFTSSTAGLRGFGGMQGGGLGYAASKHGIVGLMRTLANALAP
Mvan_3394|M.vanbaalenii_PYR-1 AGGRGGSMVFTSSTAGLRGFGGMQGGGLGYAASKHGIVGLMRALANALAP
Mflv_1904|M.gilvum_PYR-GCK AGGQGGAIVFTSSTAGLKGFGGSQGGGLGYAASKHGIVGLMRTLANALAP
MAV_3550|M.avium_104 AGNRGGVIIFTSSTAGIKGFGGLQGGGLGYAASKHGIVGLMRTLANALAP
TH_0236|M.thermoresistible__bu NGGQGGSIVITSSSAGLKGTGTDRAGGQAYAAAKRGLVGLMQVWANQLGP
MLBr_01807|M.leprae_Br4923 KQRFG-RYIFMGSVVGLSGVP----GQANYAASKSGLIGMARSLARELGS
* :: .* .*: * * *.*:* .::*: : *. *..
Mb2771|M.bovis_AF2122/97 QNIRVNSVHPCGVDTPMINNEFFQQWLT--TADMDAPHNLGNALPVELVQ
Rv2750|M.tuberculosis_H37Rv QNIRVNSVHPCGVDTPMINNEFFQQWLT--TADMDAPHNLGNALPVELVQ
MMAR_1965|M.marinum_M HSIRVNSLHPSGVDTPMINNEFIRRWLADLVAETGSGPGAGNALPVQILQ
MUL_3392|M.ulcerans_Agy99 HSIRVNSLHPSGVDPPMINNEFIRHWLADLVAETGSGPGAGNALPVQILQ
MAB_3081|M.abscessus_ATCC_1997 HNIRVNTIHPSGVATPMLINPVTEKLFADGSPEGMS--DTGNALPVVLME
MSMEG_5812|M.smegmatis_MC2_155 HRIRVNTVHPTAVNTMMAVNPAMTAFLE---NYPDGGPHLQNPMPVELLE
Mvan_3394|M.vanbaalenii_PYR-1 HNIRVNTVHPTAVNTMMAVNPAMTEFLE---HYPDGGPHLQNPMPVDLLE
Mflv_1904|M.gilvum_PYR-GCK HSIRVNSVHPTAVNTMMATNPAMTAFLE---AYPDGGPHLQNAMPVSLLE
MAV_3550|M.avium_104 LNIRVNTVHPTAVNTMMATNDDMTEFLQ---KNPGAGPHLQNPMPVGMLE
TH_0236|M.thermoresistible__bu HSIRVNTIHPTGVATGMVMNETMAKLFE---QGDEALSAMQNVLPIQILQ
MLBr_01807|M.leprae_Br4923 RSITANVVAPGFIDTEMTRAMDSKYQEA----------ALQAIPLGRIGA
* .* : * : . * :
Mb2771|M.bovis_AF2122/97 PTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
Rv2750|M.tuberculosis_H37Rv PTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
MMAR_1965|M.marinum_M ADDIAGALAWLVSDEARYITGVALPVDAGCVNKR
MUL_3392|M.ulcerans_Agy99 ADDIAGALAWLVSDEARYITGVALPVDAGCVNKR
MAB_3081|M.abscessus_ATCC_1997 PEDMANTVAWLVSEDARYITGVALPVDAGFTNRR
MSMEG_5812|M.smegmatis_MC2_155 PEDISAAIAYLVSDAAEYVTGVTFPVDAGFCNKL
Mvan_3394|M.vanbaalenii_PYR-1 PEDISAAVAYLVSDAARYVTGVTFPVDAGFCNKL
Mflv_1904|M.gilvum_PYR-GCK PEDISAAISYLVSDAAKYVTGVTFPVDAGFCNKL
MAV_3550|M.avium_104 PEDVSAAIAYLVSDEARYVTGVTFPVDAGFCNKV
TH_0236|M.thermoresistible__bu PEDIAEAVAWLVSDAGRFITGTAWPLDAGFSVR-
MLBr_01807|M.leprae_Br4923 VEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH
::: ::::*.*: . :::*.. *:*.*