For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGSLDGKVAFITGAARGQGRSHAVRLAREGANIIGIDICADIAANGYPMASRAELDQTVALVEEAGGKML GTVADVRDFGQVKAALDAGVEQFGRLDIVLANAGIAPLAFRQLSIEEELAQWRAVTGVNLDGAYHTAWAA IPHLLAGNRGGVIIFTSSTAGIKGFGGLQGGGLGYAASKHGIVGLMRTLANALAPLNIRVNTVHPTAVNT MMATNDDMTEFLQKNPGAGPHLQNPMPVGMLEPEDVSAAIAYLVSDEARYVTGVTFPVDAGFCNKV
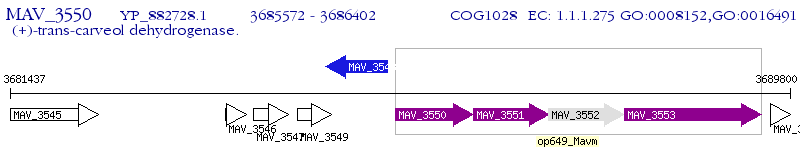
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3550 | - | - | 100% (276) | short chain dehydrogenase |
| M. avium 104 | MAV_3641 | - | 5e-75 | 53.57% (280) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. avium 104 | MAV_2623 | - | 1e-71 | 52.63% (266) | carveol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 7e-75 | 53.48% (273) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1904 | - | 1e-125 | 78.99% (276) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2750 | - | 7e-75 | 53.48% (273) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 2e-19 | 29.77% (262) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_3081 | - | 4e-69 | 51.62% (277) | Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_1965 | fabG | 3e-73 | 50.18% (277) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5812 | - | 1e-122 | 78.47% (274) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3053 | - | 4e-75 | 53.62% (276) | PUTATIVE carveol dehydrogenase |
| M. ulcerans Agy99 | MUL_3392 | fabG | 3e-72 | 49.46% (277) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_3394 | - | 1e-121 | 76.81% (276) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5812|M.smegmatis_MC2_155 -----MVS---------LDGKVAFITGAARGQGRSHAVR---LASEGADI
Mvan_3394|M.vanbaalenii_PYR-1 -----MGS---------LDGRVAFITGVARGQGRSHAVR---LAADGADI
Mflv_1904|M.gilvum_PYR-GCK -----MGS---------LDGRVAFITGVARGQGRSHAIR---LASEGAEI
MAV_3550|M.avium_104 -----MGS---------LDGKVAFITGAARGQGRSHAVR---LAREGANI
Mb2771|M.bovis_AF2122/97 ---MIDRP---------LEGKVAFITGAARGLGRAHAVR---LAADGANI
Rv2750|M.tuberculosis_H37Rv ---MIDRP---------LEGKVAFITGAARGLGRAHAVR---LAADGANI
MMAR_1965|M.marinum_M MSRPLDGP---------LSGRVAFITGAARGQGRAHALR---LARDGADV
MUL_3392|M.ulcerans_Agy99 MSRPLDGP---------LSGRVAFITGAARGQGRAHALR---LARDGADV
TH_3053|M.thermoresistible__bu VADRQPGPDLPPGRSDSLRGRVAFITGAARGQGRAHAVR---LASEGADV
MAB_3081|M.abscessus_ATCC_1997 -----MGK---------LDGKVAFITGAARGQGRAHAIK---LASEGASI
MLBr_01807|M.leprae_Br4923 ------------------MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRG
.* ...* * *:. : *. *
MSMEG_5812|M.smegmatis_MC2_155 IGVDICADIASAGYPMAGREELDETVALVEAHGGKMMSAVADVRDYGALK
Mvan_3394|M.vanbaalenii_PYR-1 IGVDICSDIASNGYPMATPAELEETVALVEAQGAKMLGSVADVRDFSAVK
Mflv_1904|M.gilvum_PYR-GCK IGVDICADIEANRYPLASRAELDETVALVESAGGKMLGSVADVRDFGALK
MAV_3550|M.avium_104 IGIDICADIAANGYPMASRAELDQTVALVEEAGGKMLGTVADVRDFGQVK
Mb2771|M.bovis_AF2122/97 IAVDICEQIASVPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLS
Rv2750|M.tuberculosis_H37Rv IAVDICEQIASVPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLS
MMAR_1965|M.marinum_M IAVDLCDQIASVPYPLGTAEELATTVKLVEDTGARIVASQADVRDREALA
MUL_3392|M.ulcerans_Agy99 IAVDLCDQIASVPYPLGTAEELATTVKLVEDTGARIVASQADVRDREALA
TH_3053|M.thermoresistible__bu IAVDLCDQIASVPYPLAGADDLAETARMVEEAGGRIVARQADVRDRDGLQ
MAB_3081|M.abscessus_ATCC_1997 IAVDICAQIPSVEYPMATTEDLDLTVKEVDALGRAISAHETDVRDREALQ
MLBr_01807|M.leprae_Br4923 IGLAVARRLVADGHRVAVT-------HRASVVPDWFFGVECDVTDNDAIG
*.: :. : : : :. .. : . ** * :
MSMEG_5812|M.smegmatis_MC2_155 TALDAGVDHFGRLDIVCANAGIAAMAFRELTIEEDLQMWADVVDVNLVGS
Mvan_3394|M.vanbaalenii_PYR-1 GALDAGVEHFGRLDIVCANAGIATMSFRELTDAEDLEMWTDVLDVNLVGT
Mflv_1904|M.gilvum_PYR-GCK SAVDAGVEHFGRLDIVCANAGIAPTAFREVSIEEDLEMWTAAIDVNLVGS
MAV_3550|M.avium_104 AALDAGVEQFGRLDIVLANAGIAPLAFRQLSIEEELAQWRAVTGVNLDGA
Mb2771|M.bovis_AF2122/97 VALQAGLDEFGRLDIVVANAGIAMMQ-------AGDDGWRDVIDVNLTGV
Rv2750|M.tuberculosis_H37Rv VALQAGLDEFGRLDIVVANAGIAMMQ-------AGDDGWRDVIDVNLTGV
MMAR_1965|M.marinum_M AALQAGIDELGQVDIVVANAGIAPMQ-------SGDDGWRDVIDVNLSGA
MUL_3392|M.ulcerans_Agy99 AALQAGIDELGQVDIVVANAGIAPMQ-------SGDDGWRDVIDVNLSGA
TH_3053|M.thermoresistible__bu QALQAGLDELGRLDIVIANAGIAPLQ-------SGPDGWRDVIDVNLTGV
MAB_3081|M.abscessus_ATCC_1997 AAFDAGVHDLGPVDIVVANAGIATLTP-----ECGDDGWRDVLDVNLTGV
MLBr_01807|M.leprae_Br4923 AAFKAVEEHQGPVEVLVANAGITSDMLIMR---MSEEQFQKVIDVNLTGV
*..* .. * :::: *****: : . .*** *
MSMEG_5812|M.smegmatis_MC2_155 FHTAKAAIPHLIAGERGGSIVFTSSTAGLRGFGGMQGGGLGYAASKHGIV
Mvan_3394|M.vanbaalenii_PYR-1 FHTAKAAIPHLIAGGRGGSMVFTSSTAGLRGFGGMQGGGLGYAASKHGIV
Mflv_1904|M.gilvum_PYR-GCK YHTAKAAIPHLIAGGQGGAIVFTSSTAGLKGFGGSQGGGLGYAASKHGIV
MAV_3550|M.avium_104 YHTAWAAIPHLLAGNRGGVIIFTSSTAGIKGFGGLQGGGLGYAASKHGIV
Mb2771|M.bovis_AF2122/97 FHTVQVAIPTLIEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVV
Rv2750|M.tuberculosis_H37Rv FHTVQVAIPTLIEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVV
MMAR_1965|M.marinum_M YYTVEVAIPTMIEQGRGGSIVLISSAAGLVGISSADAGAIGYAASKHALV
MUL_3392|M.ulcerans_Agy99 YYTVEVAIPTMIEQGRGGSIVLISSAAGLVGISSADAGAIGYVASKHALV
TH_3053|M.thermoresistible__bu HHTVEVAVPTLVEQGDGGSIVLISSVAGLVGMGGAKPGALGYTAAKHGIV
MAB_3081|M.abscessus_ATCC_1997 YHTIEVARPSMVQRGAGGSIVLTSSVAGLTGMVMDNPGGLAYTAAKHGLV
MLBr_01807|M.leprae_Br4923 FRVAKLASRNMIKQRFG-RYIFMGSVVGLSGVP----GQANYAASKSGLI
. . * :: * :: .*..*: *. * *.*:* .::
MSMEG_5812|M.smegmatis_MC2_155 GLMRTLANALAPHRIRVNTVHPTAVNTMMAVNPAMTAFLE---NYPDGGP
Mvan_3394|M.vanbaalenii_PYR-1 GLMRALANALAPHNIRVNTVHPTAVNTMMAVNPAMTEFLE---HYPDGGP
Mflv_1904|M.gilvum_PYR-GCK GLMRTLANALAPHSIRVNSVHPTAVNTMMATNPAMTAFLE---AYPDGGP
MAV_3550|M.avium_104 GLMRTLANALAPLNIRVNTVHPTAVNTMMATNDDMTEFLQ---KNPGAGP
Mb2771|M.bovis_AF2122/97 GLMRAYANHLAPQNIRVNSVHPCGVDTPMINNEFFQQWLT--TADMDAPH
Rv2750|M.tuberculosis_H37Rv GLMRAYANHLAPQNIRVNSVHPCGVDTPMINNEFFQQWLT--TADMDAPH
MMAR_1965|M.marinum_M GLMRVYANLLAPHSIRVNSLHPSGVDTPMINNEFIRRWLADLVAETGSGP
MUL_3392|M.ulcerans_Agy99 GLMRVYANLLAPHSIRVNSLHPSGVDPPMINNEFIRHWLADLVAETGSGP
TH_3053|M.thermoresistible__bu GLMRQYANLLAPHHIRVNSVHPTGVDTPMIDNEFTRQFLQQVLAEAQRPM
MAB_3081|M.abscessus_ATCC_1997 GLMRSYANMLAPHNIRVNTIHPSGVATPMLINPVTEKLFADGSPEGMS--
MLBr_01807|M.leprae_Br4923 GMARSLARELGSRSITANVVAPGFIDTEMTR-----------AMDSKYQE
*: * *. *.. * .* : * : . *
MSMEG_5812|M.smegmatis_MC2_155 HLQNPMPVELL-EPEDISAAIAYLVSDAAEYVTGVTFPVDAGFCNKL
Mvan_3394|M.vanbaalenii_PYR-1 HLQNPMPVDLL-EPEDISAAVAYLVSDAARYVTGVTFPVDAGFCNKL
Mflv_1904|M.gilvum_PYR-GCK HLQNAMPVSLL-EPEDISAAISYLVSDAAKYVTGVTFPVDAGFCNKL
MAV_3550|M.avium_104 HLQNPMPVGML-EPEDVSAAIAYLVSDEARYVTGVTFPVDAGFCNKV
Mb2771|M.bovis_AF2122/97 NLGNALPVELV-QPTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
Rv2750|M.tuberculosis_H37Rv NLGNALPVELV-QPTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
MMAR_1965|M.marinum_M GAGNALPVQIL-QADDIAGALAWLVSDEARYITGVALPVDAGCVNKR
MUL_3392|M.ulcerans_Agy99 GAGNALPVQIL-QADDIAGALAWLVSDEARYITGVALPVDAGCVNKR
TH_3053|M.thermoresistible__bu DMGNALPVQAI-EPEDVANAVAWLVSDAARYVTGVALPVDAGSVNKR
MAB_3081|M.abscessus_ATCC_1997 DTGNALPVVLM-EPEDMANTVAWLVSEDARYITGVALPVDAGFTNRR
MLBr_01807|M.leprae_Br4923 AALQAIPLGRIGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH
:.:*: : ::: ::::*.*: * *::*..:***.*