For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNPMPTHELARRMAELARQVASPRRVEDVLSDVTAEARTLIPGVDAAGVLLVGTAGRFESVAGTSELPHR LDELQMRFKEGPCLDAALDQTVVRTDDFRSEARWPQYAPAAAEIGVLSGLSFKLYTSEQAAGALNLFGLR ANAFDDEAETIGAILAAHAAAAIIAGRQSEQLESALSTRDRIGQAKGIIMERYDVDDVAAFDMLRRLSQD SNTKLTEVAAKVIETRGT
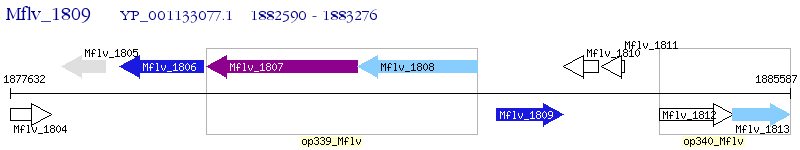
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1809 | - | - | 100% (228) | response regulator receiver/ANTAR domain-containing protein |
| M. gilvum PYR-GCK | Mflv_3842 | - | 4e-40 | 42.42% (198) | response regulator receiver/ANTAR domain-containing protein |
| M. gilvum PYR-GCK | Mflv_5218 | - | 8e-26 | 35.09% (228) | response regulator receiver/ANTAR domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2534c | - | 6e-44 | 41.20% (216) | hypothetical protein MAB_2534c |
| M. marinum M | MMAR_1808 | - | 6e-28 | 34.70% (219) | hypothetical protein MMAR_1808 |
| M. avium 104 | MAV_5172 | - | 2e-77 | 63.44% (227) | F420-dependent glucose-6-phosphate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6741 | - | 2e-80 | 64.60% (226) | ANTAR domain-containing protein |
| M. thermoresistible (build 8) | TH_2526 | - | 2e-19 | 28.77% (219) | PUTATIVE putative response regulator receiver and ANTAR domain |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4938 | - | 3e-99 | 79.39% (228) | response regulator receiver/ANTAR domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1809|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4938|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6741|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5172|M.avium_104 MTGISRRTLGRLAVGAGVLASAVQACAKPGAGHGRSGAPPAPAGKGVGFV
MAB_2534c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1808|M.marinum_M --------------------------------------------------
TH_2526|M.thermoresistible__bu --------------------------------------------------
Mflv_1809|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4938|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6741|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5172|M.avium_104 LSHEQFRTDRLVAHAQAAERAGFTHGWASDHIQPWQDNEGHSMFPWLTLA
MAB_2534c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1808|M.marinum_M --------------------------------------------------
TH_2526|M.thermoresistible__bu --------------------------------------------------
Mflv_1809|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4938|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6741|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5172|M.avium_104 LVGSSTSHVSFGTGVTCPTYRYHPATVAQAFASLAILNPGRVFLGVGTGE
MAB_2534c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1808|M.marinum_M --------------------------------------------------
TH_2526|M.thermoresistible__bu --------------------------------------------------
Mflv_1809|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4938|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6741|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5172|M.avium_104 RLNEQATTNGYGNYTERHDRLAEAIALIRQLWSGSRISFSGRYFQTNSLK
MAB_2534c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1808|M.marinum_M --------------------------------------------------
TH_2526|M.thermoresistible__bu --------------------------------------------------
Mflv_1809|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4938|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6741|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5172|M.avium_104 LYDVPATPPPIFVAAGGPKSAKLAGQFGDGWITQSGDVTNPKLLAAFGAG
MAB_2534c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1808|M.marinum_M --------------------------------------------------
TH_2526|M.thermoresistible__bu --------------------------------------------------
Mflv_1809|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4938|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6741|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5172|M.avium_104 AQAAGRDVATLGKRAEMFAVVGDDAVAARAATLWRFTAGAVDQPDPVDIQ
MAB_2534c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1808|M.marinum_M --------------------------------------------------
TH_2526|M.thermoresistible__bu --------------------------------------------------
Mflv_1809|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4938|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6741|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5172|M.avium_104 RAAESNPTDKVLGGWTGRHRRGSARRGRAAGPRRGRRSLPALSAGRPDGR
MAB_2534c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1808|M.marinum_M --------------------------------------------------
TH_2526|M.thermoresistible__bu --------------------------------------------------
Mflv_1809|M.gilvum_PYR-GCK ------------------------------------------MNPMPTHE
Mvan_4938|M.vanbaalenii_PYR-1 ------------------------------------------MTETSSHA
MSMEG_6741|M.smegmatis_MC2_155 ------------------------------------------MGESRNHE
MAV_5172|M.avium_104 HRLLPGQCLTEIALNPLRATPVSPLPATNSRRIGQNRDVMDAVDPDSRHQ
MAB_2534c|M.abscessus_ATCC_199 -----------------------------------------MTDEAAAGA
MMAR_1808|M.marinum_M ------------------------------MTATPKDSNPDSVPGGTTAD
TH_2526|M.thermoresistible__bu ------------------------------------------------MG
Mflv_1809|M.gilvum_PYR-GCK LARRMAELARQVASPRR-VEDVLSDVTAEARTLIPGVDAAGVLLVGTAGR
Mvan_4938|M.vanbaalenii_PYR-1 LAQRMAELARTVASPRR-VEDVLSDVTREVMGLIPGVDAAGVLFVGAQGR
MSMEG_6741|M.smegmatis_MC2_155 LAMRMADLARAVASPRS-LADVLTGVTTAAKELIPGVDTAGVLLIGPGNT
MAV_5172|M.avium_104 LAVRMAELVRGMAAPRR-LDQVLAEVTAAAVEVIPGADIAGVLLVRKGGE
MAB_2534c|M.abscessus_ATCC_199 FSAKIADAARRMQQEHSSVEMTLRSITTAALETVPGVEQAGITLVTGRYE
MMAR_1808|M.marinum_M PATVFAALAEIIYQGAQ-PAEMYSAICIAATLIVPGCDHA-SLLVRTNDR
TH_2526|M.thermoresistible__bu LNATVEKLIRKARASGADTCAVLTELTRSAVGDWPSIQHAGITVVEDGDV
. . : . *. : * .:
Mflv_1809|M.gilvum_PYR-GCK FESVAGTSELPHRLDELQMRFKEGPCLDAALDQTVVRTDDFRSEARWPQY
Mvan_4938|M.vanbaalenii_PYR-1 FESVAGTSELPHRLDELQMKYRQGPCLEAALDSLIVRTDDFRTEQRWPLY
MSMEG_6741|M.smegmatis_MC2_155 FESLAALDELPHALDELQMRFDEGPCRQAALADLVVRTEDFRAEQRWPRY
MAV_5172|M.avium_104 FETLADTDSLAARLDVLQHDFGEGPCAQAALQETIVRSDDLRREPRWPCY
MAB_2534c|M.abscessus_ATCC_199 IESRAATSETPRKLDDIQQELREGPCVQSAWEHETVYAEDLAQTTPWPRF
MMAR_1808|M.marinum_M YITVGASDRLAQHVDELERKAGDGPCIDAIEEETPQIETDLTTPTQWPKF
TH_2526|M.thermoresistible__bu VRSIAATDGHVLVVDNIARRLSTGPGIDAALDEDSYRIDDLRADSRWPLF
: . . :* : ** :: *: ** :
Mflv_1809|M.gilvum_PYR-GCK APAAAEIG-VLSGLSFKLYTSEQA------------AGALNLFGLRANAF
Mvan_4938|M.vanbaalenii_PYR-1 SAAAAELG-VLSGLSFKLYTSART------------AGALNLFALKPNAF
MSMEG_6741|M.smegmatis_MC2_155 SPASVELG-VLSGLSFKLYTADRT------------AGALNLFGFSPMNW
MAV_5172|M.avium_104 APAAVQLG-VLSSLSFKLYTADRT------------AGALNLFSHRPDAW
MAB_2534c|M.abscessus_ATCC_199 AEAAVKLG-VRSMLSFQLFTFGEN------------LGALNLYAQSPGAF
MMAR_1808|M.marinum_M AAALLAQTPVRGAMGFRLLVDKRK------------GAALNLFSETPNSF
TH_2526|M.thermoresistible__bu ADKVAASTPIRSVIAIPVGNPIRNPIRNPIASAGTVRAALSLYSDLPGAF
: : . :.: : . .**.*:. . :
Mflv_1809|M.gilvum_PYR-GCK DDEAETIGAILAAHAAAAIIAGRQ--------SEQLESALSTRDRIGQAK
Mvan_4938|M.vanbaalenii_PYR-1 DADAETIGAVFAAHAAAAIMASRE--------NEQLQSALSTRDRIGQAK
MSMEG_6741|M.smegmatis_MC2_155 DEDAVTTGTVLAAHAASAIVASRQ--------EEQLKTALLSRDRIGQAK
MAV_5172|M.avium_104 DTEAETIGSVFAAHAAAAILAGSR--------AEQLYSAVSTRDRIGQAK
MAB_2534c|M.abscessus_ATCC_199 GADSYDAGLALATHAAIVLVAAQR--------ESQWASALASRDVIGQAK
MMAR_1808|M.marinum_M DTESAGRAAVLASFASVAVNAIAHG-----EDAASLRRGLLSNREIGKAV
TH_2526|M.thermoresistible__bu DDEVEAVAATYAEHIGRVLTARRGRRRAPVSGSARAQGSVRGQDAIETSK
. : . * . . .: * .: . * :
Mflv_1809|M.gilvum_PYR-GCK GIIMERYDVDDVAAFDMLRRLSQDSNTKLTEVAAKVIETRGT--------
Mvan_4938|M.vanbaalenii_PYR-1 GIIMERYDVDDVTAFEMLRRLSQDSNTKLVEVAAKVIETRGT--------
MSMEG_6741|M.smegmatis_MC2_155 GIIMERYKVDDVQAFEMMRQLSQHSNTRLVDIAGQIIESR----------
MAV_5172|M.avium_104 GIIMERFGVDDVRAFDLLRRLSQESQVKLVEIAQQIIDTRGQGA------
MAB_2534c|M.abscessus_ATCC_199 GMLMERFSINAAEAFTVIRTLSQDFNTKLVEIARAIVEDRPLG-------
MMAR_1808|M.marinum_M GMLMLLNGFTEEEAFDLLRHHSQSLNIKLADVARTVIENRGQLPPEALET
TH_2526|M.thermoresistible__bu AQLMKHFGVDPAAALTVLLRLSRECNRSLEDVARLLVGPRPVAEPPSQAT
. :* . *: :: *: : * ::* :: *
Mflv_1809|M.gilvum_PYR-GCK --------------------
Mvan_4938|M.vanbaalenii_PYR-1 --------------------
MSMEG_6741|M.smegmatis_MC2_155 --------------------
MAV_5172|M.avium_104 --------------------
MAB_2534c|M.abscessus_ATCC_199 --------------------
MMAR_1808|M.marinum_M DDTVNPTVSG----------
TH_2526|M.thermoresistible__bu ATATPDHAGGRPPVWAEGAA