For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLLSLVDRTVLVTGGGSGIGKGVAAAVVASGGNVVLAGRNADKLAAAAEEITAQSGDGAGEVMYEPADVT DEDAVTRLVQAATGWRGRLDAVVHSAGGSETIGPLTQVDSAAWRNTVDLNVNGTMYVLKHAAREMVRGGG GAFIGISSIASSNTHRWFGAYGPSKAALDHLMMVAADELGPSSVRVNGIRPGLTKTDMVAPILDTPAVSD DYAACTPLPRVGEVEDIANLAVFLLSDAASFITGQVINVDGGHGLRRGPDLSGMLEPLFGADGLRGVVAE
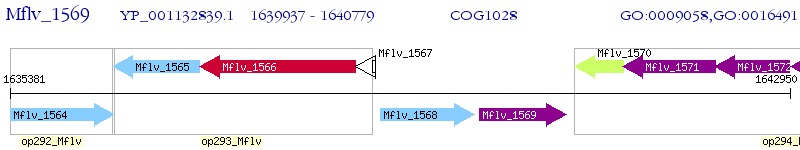
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1569 | - | - | 100% (280) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2071 | - | 2e-41 | 39.25% (265) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_1202 | - | 7e-32 | 35.06% (251) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3515c | - | 1e-116 | 72.66% (278) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv3485c | - | 1e-116 | 72.66% (278) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 4e-24 | 30.47% (256) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0952 | - | 2e-30 | 35.47% (265) | short chain dehydrogenase |
| M. marinum M | MMAR_4968 | - | 1e-113 | 71.38% (276) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0671 | - | 1e-108 | 67.97% (281) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5885 | - | 1e-125 | 78.21% (280) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1273 | - | 1e-99 | 68.13% (273) | PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE |
| M. ulcerans Agy99 | MUL_4042 | - | 1e-112 | 71.01% (276) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5189 | - | 1e-145 | 90.68% (279) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3515c|M.bovis_AF2122/97 --------------------------------------------------
Rv3485c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4968|M.marinum_M --------------------------------------------------
MUL_4042|M.ulcerans_Agy99 --------------------------------------------------
MAV_0671|M.avium_104 --------------------------------------------------
Mflv_1569|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5189|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5885|M.smegmatis_MC2_155 --------------------------------------------------
TH_1273|M.thermoresistible__bu MGWSAVAKPDASEQVTYETLDDGRIARIWLNRPEAQNAQSRTLLVQLDEA
MLBr_01807|M.leprae_Br4923 --------------------------------------------------
MAB_0952|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3515c|M.bovis_AF2122/97 --------------------------------------------------
Rv3485c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4968|M.marinum_M --------------------------------------------------
MUL_4042|M.ulcerans_Agy99 --------------------------------------------------
MAV_0671|M.avium_104 --------------------------------------------------
Mflv_1569|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5189|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5885|M.smegmatis_MC2_155 --------------------------------------------------
TH_1273|M.thermoresistible__bu FLRAEADDQVRVVILAARGRNFSAGHDLGSEEALLERQPGPDQHPTFRSH
MLBr_01807|M.leprae_Br4923 --------------------------------------------------
MAB_0952|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3515c|M.bovis_AF2122/97 --------------------------------------------------
Rv3485c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4968|M.marinum_M --------------------------------------------------
MUL_4042|M.ulcerans_Agy99 --------------------------------------------------
MAV_0671|M.avium_104 --------------------------------------------------
Mflv_1569|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5189|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5885|M.smegmatis_MC2_155 --------------------------------------------------
TH_1273|M.thermoresistible__bu GATATGIAERTYRQEWHFYFENTCRWRDLRKITIAQVQGNAISAALMLIW
MLBr_01807|M.leprae_Br4923 --------------------------------------------------
MAB_0952|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3515c|M.bovis_AF2122/97 ----MNSRAPRNLAVSSPSAQVTGRMVQNGENLFQFRREGPQVQLS----
Rv3485c|M.tuberculosis_H37Rv ----MNSRAPRNLAVSSPSAQVTGRMVQNGENLFQFRREGPQVQLS----
MMAR_4968|M.marinum_M ------------------------------------------MQLS----
MUL_4042|M.ulcerans_Agy99 ------------------------------------------MQLS----
MAV_0671|M.avium_104 ------------------------------------------MQLS----
Mflv_1569|M.gilvum_PYR-GCK ------------------------------------------MLLS----
Mvan_5189|M.vanbaalenii_PYR-1 ------------------------------------------MLLS----
MSMEG_5885|M.smegmatis_MC2_155 ------------------------------------------MQLS----
TH_1273|M.thermoresistible__bu ACDLIVAADDARFSDVVGVRMGMPGVEYYAHPWEFGPRKAKELLLTGDSI
MLBr_01807|M.leprae_Br4923 ----------------------------MTDAAAKVSAAPGRGKPA----
MAB_0952|M.abscessus_ATCC_1997 ------------------------------MVVESAEPAERKVMFD----
Mb3515c|M.bovis_AF2122/97 --------------------------------FQDR----TYLVTGGG--
Rv3485c|M.tuberculosis_H37Rv --------------------------------FQDR----TYLVTGGG--
MMAR_4968|M.marinum_M --------------------------------FQDR----TYLVTGGG--
MUL_4042|M.ulcerans_Agy99 --------------------------------FQDR----TYLVTGGG--
MAV_0671|M.avium_104 --------------------------------FQDR----TYLITGGG--
Mflv_1569|M.gilvum_PYR-GCK --------------------------------LVDR----TVLVTGGG--
Mvan_5189|M.vanbaalenii_PYR-1 --------------------------------LSER----TYLVTGGG--
MSMEG_5885|M.smegmatis_MC2_155 --------------------------------LAER----TYLVTGGG--
TH_1273|M.thermoresistible__bu DADEAYRMGMVSKVFPRAELEEKTLEFAR--RIAERPTMAALLIKDSGNA
MLBr_01807|M.leprae_Br4923 ---------------------------FVSR---------SILVTGGN--
MAB_0952|M.abscessus_ATCC_1997 ---------------------------LSAR---------SVLVTGGT--
: *:...
Mb3515c|M.bovis_AF2122/97 --SGIGKGVAAGLVAAG-----AAVMIVG---RNPDKLAAAVKDIEALKT
Rv3485c|M.tuberculosis_H37Rv --SGIGKGVAAGLVAAG-----AAVMIVG---RNPDKLAAAVKDIEALKT
MMAR_4968|M.marinum_M --SGIGKGVAAGLVAAG-----ASVMIVG---RNPDKLAGAVQELEALGA
MUL_4042|M.ulcerans_Agy99 --SGIGKGVAAGLVAAG-----ASVMIVG---RNPDKLAGAVQELEALGA
MAV_0671|M.avium_104 --SGIGKGVAAGLVSAG-----ASVMIVG---RNPDRLAGAVEEIAPLAD
Mflv_1569|M.gilvum_PYR-GCK --SGIGKGVAAAVVASG-----GNVVLAG---RNADKLAAAAEEITAQSG
Mvan_5189|M.vanbaalenii_PYR-1 --SGIGKGVAAAVVASG-----GNAVLAG---RNADKLAAAAEEIKAQTG
MSMEG_5885|M.smegmatis_MC2_155 --SGIGKGAAAAIVAAG-----GDVMLVG---RNADKLSAAADEIGAL--
TH_1273|M.thermoresistible__bu ASDAMGXXXQPPVPAGGRLDRLRDLVLIGDVGRMGAHLAARPGEFACGGG
MLBr_01807|M.leprae_Br4923 --RGIGLAVARRLVADG-----HRVAVTH---RAS------------VVP
MAB_0952|M.abscessus_ATCC_1997 --KGIGRGIATVFARAG-----ANVAVAA---RSPRELSSVTAELGELGA
.:* . * : *
Mb3515c|M.bovis_AF2122/97 -----GAIGYEPADITDEEQ------------------------------
Rv3485c|M.tuberculosis_H37Rv -----GAIGYEPADITDEEQ------------------------------
MMAR_4968|M.marinum_M NG---GAIRYEPTDITNEDE------------------------------
MUL_4042|M.ulcerans_Agy99 NG---GAIRYEPTDITNEDE------------------------------
MAV_0671|M.avium_104 RAGNGGAIRYEPTDVTNEDE------------------------------
Mflv_1569|M.gilvum_PYR-GCK D--GAGEVMYEPADVTDEDA------------------------------
Mvan_5189|M.vanbaalenii_PYR-1 D--GAGEVIYEPADVTDEDQ------------------------------
MSMEG_5885|M.smegmatis_MC2_155 ---GPGSVRYEPADVTDEDE------------------------------
TH_1273|M.thermoresistible__bu Q--PVGIASDDQHLATGRGDRGGHPFADTASAAGHQIRPVGQRQLHGHPP
MLBr_01807|M.leprae_Br4923 DW-----FFGVECDVTDNDA------------------------------
MAB_0952|M.abscessus_ATCC_1997 GN-----VIGVRLDVSDPGS------------------------------
:.
Mb3515c|M.bovis_AF2122/97 -TLRVVDAA--TAWHGRLHGVVHCAGGSQTIGPITQIDSQAWRRTVDLNV
Rv3485c|M.tuberculosis_H37Rv -TLRVVDAA--TAWHGRLHGVVHCAGGSQTIGPITQIDSQAWRRTVDLNV
MMAR_4968|M.marinum_M -TARAVDAV--TAWHGRLHGVVHCAGGSETIGPITQVDSEAWRRTVDLNV
MUL_4042|M.ulcerans_Agy99 -TARAVDAV--TAWHGRLHGVVHCAGGSENIGPITQVDSEAWRRTVDLNV
MAV_0671|M.avium_104 -VVRAVDAA--TAWHGRLHGAVHCAGGSLTVGPITHTDSEAWRNTVDLNV
Mflv_1569|M.gilvum_PYR-GCK -VTRLVQAA--TGWRGRLDAVVHSAGGSETIGPLTQVDSAAWRNTVDLNV
Mvan_5189|M.vanbaalenii_PYR-1 -AARLVEAA--TAWHGRLDGVVHSAGGSETIGPITQIDSAAWRNTVDLNV
MSMEG_5885|M.smegmatis_MC2_155 -VHRAVDAA--TAWHGRLHGVVHSAGGSQTIGPITQIDSELWRQTIDLNI
TH_1273|M.thermoresistible__bu FRSRDITAGGLXGWHARLHGVVHCAGGSETIGPITQVDSEAWRRTVDLNV
MLBr_01807|M.leprae_Br4923 ----IGAAFKAVEEHQGPVEVLVANAGITSDMLIMRMSEEQFQKVIDVNL
MAB_0952|M.abscessus_ATCC_1997 ----CADAARTVVDAFGALDVVCANAGIFPEARLDTMTPEQLSEVLDVNV
* .: . .* : ..:*:*:
Mb3515c|M.bovis_AF2122/97 NGTMYVLKHAARELVRGGGGSFVGISSIAASNTHR-WFGAYGVTKSAVDH
Rv3485c|M.tuberculosis_H37Rv NGTMYVLKHAARELVRGGGGSFVGISSIAASNTHR-WFGAYGVTKSAVDH
MMAR_4968|M.marinum_M NGTMYVLKHAAREMVRGGGGSFVGISSIAASNTHR-WFGAYGVTKSAVDH
MUL_4042|M.ulcerans_Agy99 NGTMYVLKHAAREMVRGGGGSFVGISSIAASNTHR-WFGAYGVTKSAVDH
MAV_0671|M.avium_104 NGTMYVLKHVGRELVRGGGGSFIGISSIAASNTHR-WFGPYGVTKSAIDH
Mflv_1569|M.gilvum_PYR-GCK NGTMYVLKHAAREMVRGGGGAFIGISSIASSNTHR-WFGAYGPSKAALDH
Mvan_5189|M.vanbaalenii_PYR-1 NGTMYVLKHSAREMVRGGGGSFIGISSIASSNTHR-WFGAYGPSKAALDH
MSMEG_5885|M.smegmatis_MC2_155 NGTMYLLKHAGREMVRGGGGSFVGISSIASSNIHR-WFGAYGVSKAALDH
TH_1273|M.thermoresistible__bu NGTMYLIKHAAREMVRGGGGSFVGVSSIASSNTHR-WFGAYGPSKAALDH
MLBr_01807|M.leprae_Br4923 TGVFRVAKLASRNMIKQRFGRYIFMGSVVG-LSGVPGQANYAASKSGLIG
MAB_0952|M.abscessus_ATCC_1997 KGTVYTVQACLAPLTASGRGRVILTSSITGPVTGYPGWSHYGASKAAQLG
.*.. : : * : .*:.. . *. :*:.
Mb3515c|M.bovis_AF2122/97 MMKLAADELGPSWVRVNSIRPGLIRTDLVVP-VTESPELSADYRVCTPLP
Rv3485c|M.tuberculosis_H37Rv MMKLAADELGPSWVRVNSIRPGLIRTDLVVP-VTESPELSADYRVCTPLP
MMAR_4968|M.marinum_M LMQLAADELGASWVRVNSIRPGLIRTDLVAA-ITESAELSSDYAMCTPLP
MUL_4042|M.ulcerans_Agy99 LMQLAADELGASWVRVNSIRPGLIRTDLVAA-ITESAELSSDYAMCTPLP
MAV_0671|M.avium_104 MMMLAADELGESWVRVNSIRPGLIRTDLVDASVIQSPEISADYAQCTPLP
Mflv_1569|M.gilvum_PYR-GCK LMMVAADELGPSSVRVNGIRPGLTKTDMVAP-ILDTPAVSDDYAACTPLP
Mvan_5189|M.vanbaalenii_PYR-1 LMMVAADELGPSSVRVNGIRPGLTRTDMVAP-IVDTPALSEDYAACTPLP
MSMEG_5885|M.smegmatis_MC2_155 LLKLAADELGPSWVRVNSIRPGLTRTEMVEP-ILGVPAVTDDYAACTPLP
TH_1273|M.thermoresistible__bu LLMVAADELGPSWVRVNGIRPGLTKTDLVAP-IMDTPAVVDDYRACTPLP
MLBr_01807|M.leprae_Br4923 MARSLARELGSRSITANVVAPGFIDTEMTRA---MDSKYQEAALQAIPLG
MAB_0952|M.abscessus_ATCC_1997 FMRTAAIELAPRGVTVNAILPGNILTEGLVD---MGEEYISGMARSIPMG
: * **. : .* : ** *: . *:
Mb3515c|M.bovis_AF2122/97 RVGEVEDVANLAMFLLSDAASWITGQVINVDGGHMLRRGPDFSPMLEPVF
Rv3485c|M.tuberculosis_H37Rv RVGEVEDVANLAMFLLSDAASWITGQVINVDGGHMLRRGPDFSPMLEPVF
MMAR_4968|M.marinum_M RQGEVEDVANMAMFLLSDAASFVTGQVINVDGGQMLRRGPDFSAMLEPVF
MUL_4042|M.ulcerans_Agy99 RQGEVEDVANMAMFLLSDAASFVTGQVINVDGGQMLRRGPDFSAMLEPVF
MAV_0671|M.avium_104 RVGEVEDVANLAMFLLSDAAGWITGQCINVDGGHMLRRGPDYSSMMVQMF
Mflv_1569|M.gilvum_PYR-GCK RVGEVEDIANLAVFLLSDAASFITGQVINVDGGHGLRRGPDLSGMLEPLF
Mvan_5189|M.vanbaalenii_PYR-1 RVGEVEDIANLAVFLLSDAASFITGQVINVDGGHGLRRGPDLSAMLEPIF
MSMEG_5885|M.smegmatis_MC2_155 RFGEVEDIANLAVFLLSDAAGWITGQAINVDGGHGLRRGPDISGMLEPLF
TH_1273|M.thermoresistible__bu RFGEVSDIANLAVFLLSDAASWITGQIINVDGGHGLRRGPDMSGMLEPVF
MLBr_01807|M.leprae_Br4923 RIGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH------------
MAB_0952|M.abscessus_ATCC_1997 MLGSPVDIGHLAAFLATDEAGYITGQAIVVDGGQVLPESPDAVNP-----
* ::. . ** :: *.:::* * **** : .
Mb3515c|M.bovis_AF2122/97 GADGLRGVVG--
Rv3485c|M.tuberculosis_H37Rv GADGLRGVVG--
MMAR_4968|M.marinum_M GRDALRGLV---
MUL_4042|M.ulcerans_Agy99 GRDALRGLV---
MAV_0671|M.avium_104 GQDALRGVV---
Mflv_1569|M.gilvum_PYR-GCK GADGLRGVVAE-
Mvan_5189|M.vanbaalenii_PYR-1 GADGLRGVVST-
MSMEG_5885|M.smegmatis_MC2_155 GAEGLRGVVTDG
TH_1273|M.thermoresistible__bu GADGLRGVVEG-
MLBr_01807|M.leprae_Br4923 ------------
MAB_0952|M.abscessus_ATCC_1997 ------------