For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NDRLLDGLASSIVERGYRETTVADIVRHAKTSKRAFYEQFASKEQCLIELLRNNNTDLIAHINAAVDPES DWREQIGQAVDAYIGHISARPAITLCWIREAPALGAVARPLHRQVMRDLTEMLTALTGSSGFRRAGLQPI EPAIALILLGGLRELTALFVEDGRDMRGIAGPATAAAVALLGPH*
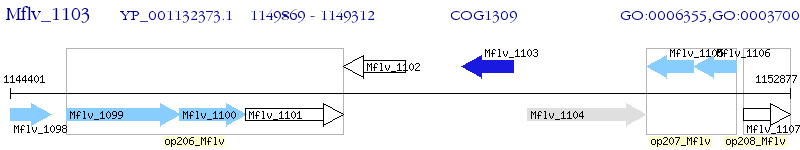
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1103 | - | - | 100% (185) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1674 | - | 3e-47 | 51.10% (182) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_5172 | - | 2e-08 | 31.25% (112) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0140c | - | 1e-59 | 61.88% (181) | transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv0135c | - | 1e-59 | 61.88% (181) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_02568 | - | 3e-06 | 23.57% (140) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1149 | - | 3e-06 | 25.00% (164) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0345 | - | 6e-59 | 61.33% (181) | transcriptional regulatory protein |
| M. avium 104 | MAV_5161 | - | 1e-58 | 62.98% (181) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6480 | - | 7e-60 | 62.57% (179) | putative transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_0083 | - | 2e-63 | 65.92% (179) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_4775 | - | 5e-59 | 61.33% (181) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5713 | - | 4e-74 | 75.00% (180) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1103|M.gilvum_PYR-GCK ----------------------MNDRLLDGLASSIVERGYRETTVADIVR
Mvan_5713|M.vanbaalenii_PYR-1 ----MRTTPLAAGPHP-----SVADRLLDGLASAITERGYRDSTVADVVR
MSMEG_6480|M.smegmatis_MC2_155 ----MTTALTGTATAGGSGGDAFRSRLLDGLSTAIDDKGYRDSTVADIVR
TH_0083|M.thermoresistible__bu ----MTTAVQ-PADVG-AGDDAFRGRLLSGLAASIAERGYRETTVADIVR
Mb0140c|M.bovis_AF2122/97 ----MTAVAAGALVVE---TDSFRLRLLDGLVASIGERGYRATTVSDIVR
Rv0135c|M.tuberculosis_H37Rv ----MTAVAAGALVVE---TDSFRLRLLDGLVASIGERGYRATTVSDIVR
MMAR_0345|M.marinum_M ----MTAAASSPAALD---SDPFRQRLLEGLVASIGERGYHATTVADIVR
MUL_4775|M.ulcerans_Agy99 ----MTAAASSPAALD---SDPFRQRLLEGLVASIGERGYHATTVADIVR
MAV_5161|M.avium_104 ----MTATDGEPDLGE---IDPFRLRLLDGLATSIGERGYRASTVADVVR
MAB_1149|M.abscessus_ATCC_1997 MAVPSRPDVPGRTPRVRMAGSERRRQLIDVARSLFAERGYEATSIEEIAQ
MLBr_02568|M.leprae_Br4923 -----MAGGAKRIPRA-----VREQQMLDAAVQMFSVNGYHKTSMDAIAA
:::. : .**. ::: :.
Mflv_1103|M.gilvum_PYR-GCK HAKTSKRAFYEQFASKEQCLIELLRNNNTDLIAHINAAVDPESDWREQIG
Mvan_5713|M.vanbaalenii_PYR-1 YARTSKRTFYEQFASKEECLIEMLRRNHSDLIAGIGAAVEPEADWRLQID
MSMEG_6480|M.smegmatis_MC2_155 HARTSKRTFYDQFASKEACFIELLRANNAALVEHIRSAVSPEAEWQDQID
TH_0083|M.thermoresistible__bu HARTSKRTFYDQFGSKEECFQELLWANNIDMIGAIRAAVDPEADWSEQIR
Mb0140c|M.bovis_AF2122/97 HARTSKRTFYDRFTSKEQCFLELLLADNETLGNSIRAAVDPNADWHDQIR
Rv0135c|M.tuberculosis_H37Rv HARTSKRTFYDRFTSKEQCFLELLLADNETLGNSIRAAVDPNADWHDQIR
MMAR_0345|M.marinum_M HARTSKRTFYDRFESKEQCFLDLLRADNERLAKRIRAAVDPEADWQAQIR
MUL_4775|M.ulcerans_Agy99 HARTSKRTFYDRFESKEQCFLDLLRADNERLAKRIRAAVDPEADWQAQIR
MAV_5161|M.avium_104 CARTSKRTFYDHFAGKEECFLELLAVDIEKLGAKIAASVDPEADWHVQIR
MAB_1149|M.abscessus_ATCC_1997 RAGVSKPVVYEHFGGKEGLYAVVVDREMSVLLDGITSSLTS-YRSRVRLE
MLBr_02568|M.leprae_Br4923 EAQISKPMLYLYYGSKEDLFGACLNREMSRFIAMVRADIDFTQSPKDLLR
* ** .* : .** : : : : : : :
Mflv_1103|M.gilvum_PYR-GCK QAVDAYIGHISARPAITLCWIREAPALGAVARPLHRQVMRDLTEMLTALT
Mvan_5713|M.vanbaalenii_PYR-1 KAVGAYIDHISARPAITLCWIREAPGLGAAARPLHRLVMGDLTDLLVSLT
MSMEG_6480|M.smegmatis_MC2_155 QAVGAYVEHIESAPSITLSWIRELPALGPAARPLQRDAMGALTDLLIDLS
TH_0083|M.thermoresistible__bu QAVVAYVDHIESRPAITLAWIRELPAIGASVRPIQRAMMHELTELLIELS
Mb0140c|M.bovis_AF2122/97 QAVEAYVTHIESRPAVTLSWIREFPSLGAAAYPVQRRGMEQLTSLLIELS
Rv0135c|M.tuberculosis_H37Rv QAVEAYVTHIESRPAVTLSWIREFPSLGAAAYPVQRRGMEQLTSLLIELS
MMAR_0345|M.marinum_M QAVEAYVSHIESRLAVTLSWIREFPSLGPISYPVQRHGMNLLTSLLVELS
MUL_4775|M.ulcerans_Agy99 QAVEAYLSHIESRLAVTLSWIREFPSLGPISYPVQRHGMNLLTSLLVELS
MAV_5161|M.avium_104 QAVEAYVGYIEARPAITLSWIRELPSLGAAARPVQRRGLQLLTSLLIDLS
MAB_1149|M.abscessus_ATCC_1997 QVALALLSYIEERTDGFRILIRDSP-PGVSSGTYSSLLNDAVSQVSSILA
MLBr_02568|M.leprae_Br4923 NTIVSFLRYIDANRASWIVMYIQAMSLPAFAQTVR-EARDQITELVAGLM
:. : : :*. : . ::.: *
Mflv_1103|M.gilvum_PYR-GCK GSSGFRRAGLQPIEPAIALILLGGLRELTALFVEDGRDMRGIAGPATAAA
Mvan_5713|M.vanbaalenii_PYR-1 GNPGFRRAGLDPITAPVALILVGGLRELTALFVEDERDVHGIAAPAVAAA
MSMEG_6480|M.smegmatis_MC2_155 DSPGFRRAGLNGLSRQMAIILLGGLRELTALIVEDGHDIREIYEPAVAAS
TH_0083|M.thermoresistible__bu RSPGFARAQMPPLTRQLAVFLLGGLRELTALFVEDGRDVRGLIAPAVSAS
Mb0140c|M.bovis_AF2122/97 ASPGFRRANLPPLNVPLAVILLGGLRELTALTVEDGQPIRNIVEPAVDAS
Rv0135c|M.tuberculosis_H37Rv ASPGFRRANLPPLNVPLAVILLGGLRELTALTVEDGQPIRNIVEPAVDAS
MMAR_0345|M.marinum_M ASPGFRRAGLPAVPGALAVILLGGLRELTALTVEDGRDIREIVEPAVDAS
MUL_4775|M.ulcerans_Agy99 ASPGFRRAGLPAVPGALAVILLGGLRELTALTVEDGRDIREIVEPAVDAS
MAV_5161|M.avium_104 ASPGFRRAQLPPLTAPLAVILLGGLRELTALAVEDGKPVRGIVEPAVDAS
MAB_1149|M.abscessus_ATCC_1997 G--DFQRRGLDAEMAPLYAQALVGQVSMTAQWWLDAREPKKEVVAAHIVN
MLBr_02568|M.leprae_Br4923 R--VGTRSSRPDHEYEIMAGALVGAGEAMATRLTTG-DIAVDEAAELMIN
* : : * . * .
Mflv_1103|M.gilvum_PYR-GCK VALLGPH---------------
Mvan_5713|M.vanbaalenii_PYR-1 TALVSAG---------------
MSMEG_6480|M.smegmatis_MC2_155 RAILTASSAEHQAR--------
TH_0083|M.thermoresistible__bu KALLSAGASE------------
Mb0140c|M.bovis_AF2122/97 IALLGPRS--------------
Rv0135c|M.tuberculosis_H37Rv IALLGPRS--------------
MMAR_0345|M.marinum_M IALVGPRH--------------
MUL_4775|M.ulcerans_Agy99 IALVGPRH--------------
MAV_5161|M.avium_104 VALLGPRH--------------
MAB_1149|M.abscessus_ATCC_1997 LCWHGLKHLEADPQLHEE----
MLBr_02568|M.leprae_Br4923 LFWHGLKGAPADRDFGPSIVAG