For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVALIAPHHFPIREPFAGGIESHVWHLARALIRQGHAVTLCAAAGSDLAMEHPALTVTAMPRSAAASRVF PPPGAIKESAHHAFVELMSELADPASGVDVVHNHSLNPVPVLVARRLPVPVLTTLHTPPFPALEAAVGAV ARSGQAFAAVSRQAAAAWEHVGIRRMALVRNGIDTSRWPAGPGGEDLVWSGRLLEEKGPHLAVDAALRAG RRIVLAGPIVDDHFFRRRIAPTLGSRASYAGHLGQRELATLVGRAAAALVTPLWDEPYGQVVVEAMACGT PVVAFAGGGIPELVDEHSGRLVEPGDVAALAAAVREAVALPRAGVRRVAVQRHGLARMVTGYVALYRALI AEATPPPRPPKLHSSSESANQSQAVSATARACLTNVSGVSAET*
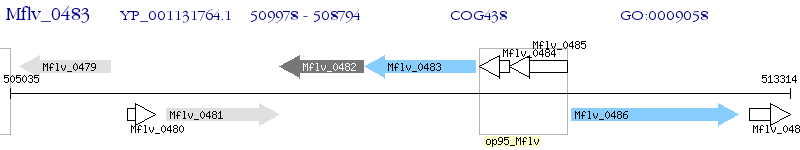
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0483 | - | - | 100% (394) | glycosyl transferase, group 1 |
| M. gilvum PYR-GCK | Mflv_4262 | - | 1e-21 | 27.59% (406) | glycosyl transferase, group 1 |
| M. gilvum PYR-GCK | Mflv_3449 | - | 4e-18 | 28.73% (369) | glycosyl transferase, group 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3058 | - | 6e-25 | 28.71% (418) | transferase |
| M. tuberculosis H37Rv | Rv3032 | - | 6e-25 | 28.71% (418) | transferase |
| M. leprae Br4923 | MLBr_01715 | - | 9e-21 | 28.18% (369) | putative transferase |
| M. abscessus ATCC 19977 | MAB_3366 | - | 2e-16 | 27.53% (385) | glycosyltransferase |
| M. marinum M | MMAR_1681 | - | 2e-23 | 29.81% (369) | transferase |
| M. avium 104 | MAV_1024 | - | 5e-32 | 30.05% (396) | glycosyl transferase, group 1 family protein |
| M. smegmatis MC2 155 | MSMEG_5641 | - | 4e-32 | 30.19% (361) | glycosyl transferase, group 1 family protein |
| M. thermoresistible (build 8) | TH_0068 | - | 3e-39 | 32.09% (374) | glycosyl transferase, group 1 family protein |
| M. ulcerans Agy99 | MUL_1919 | - | 1e-23 | 29.81% (369) | transferase |
| M. vanbaalenii PYR-1 | Mvan_2096 | - | 4e-23 | 29.32% (399) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3058|M.bovis_AF2122/97 ------------------------MRILMVS---WEYPPVVIGGLGRHVH
Rv3032|M.tuberculosis_H37Rv ------------------------MRILMVS---WEYPPVVIGGLGRHVH
MLBr_01715|M.leprae_Br4923 MFIQRHAVVRGIFGTFAPAEGTTLMKILIVS---WEYPPVVIGGLGRHVH
MMAR_1681|M.marinum_M ------------------------MKILMVS---WEYPPVVIGGLGRHVH
MUL_1919|M.ulcerans_Agy99 ------------------------MKILMVS---WEYPPVVIGGLGRHVH
Mvan_2096|M.vanbaalenii_PYR-1 ----------------------MKTKILLVS---WEYPPVVIGGLGRHVH
MAB_3366|M.abscessus_ATCC_1997 ------------------------MRILMVS---WEYPPVVIGGLGRHVH
MAV_1024|M.avium_104 ------MRGADFVLPTDVPEAGRPLRIVLVAPPYFDVPPDGYGGVEAVVA
MSMEG_5641|M.smegmatis_MC2_155 ------MSGNDILAPADVDD---PLRIVLVAPPYFDIPPKGYGGTEAVVA
TH_0068|M.thermoresistible__bu -----------VARPAHQLG--RRLRVALVAPPYFDVPPRAYGGIEAVVA
Mflv_0483|M.gilvum_PYR-GCK ------------------------MRVALIAPHHFPIREPFAGGIESHVW
:: ::: : ** *
Mb3058|M.bovis_AF2122/97 HLSTALAAAGHDVVVLSRCPSGTDPSTHPSSDEVTEGVRVIAAAQDPHEF
Rv3032|M.tuberculosis_H37Rv HLSTALAAAGHDVVVLSRCPSGTDPSTHPSSDEVTEGVRVIAAAQDPHEF
MLBr_01715|M.leprae_Br4923 HLSTALAAAGHDVVVLSRRPSGTDPCTHPTSDEISEGVRVIAAAQDPHEF
MMAR_1681|M.marinum_M HLATALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVIAAAQDPHEF
MUL_1919|M.ulcerans_Agy99 HLATALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVIAAAQDPHEF
Mvan_2096|M.vanbaalenii_PYR-1 HLATELAQSGHEVVVLSRRPTATDPQTHPTTDEVHEGVRVIAAAEDPHEF
MAB_3366|M.abscessus_ATCC_1997 HLSTELAAAGHDVVVLTRRPSGTDPSSHPTSDEISEGVRVVAAAEDPHDF
MAV_1024|M.avium_104 DLADMLVARGHQVTLLGAGEPGTAARFIPLWDRTVPERLGEPYPEIMHAL
MSMEG_5641|M.smegmatis_MC2_155 DLADTLTARGHHVTVLGAGAPGTAARFQPLWERTLPDRLGQPYPEIMHAL
TH_0068|M.thermoresistible__bu DLADALVAHGHEVTLIGAGEPGTSARFEQVWERSVPELLGKPLPEMIHAL
Mflv_0483|M.gilvum_PYR-GCK HLARALIRQGHAVTLCAAAGSDLAMEHPALTVTAMPRSAAASRVFPPPGA
.*: * ** *.: . .
Mb3058|M.bovis_AF2122/97 TFGNDMMAWTLAMGHAMIRAGLRLKKLGTDRSWRPDVVHAHDWLVAHPAI
Rv3032|M.tuberculosis_H37Rv TFGNDMMAWTLAMGHAMIRAGLRLKKLGTDRSWRPDVVHAHDWLVAHPAI
MLBr_01715|M.leprae_Br4923 TFSNDMMAWTLAMGHAMIRTGLSLTRHSSDLPWRPDVVHAHDWLVAHPAI
MMAR_1681|M.marinum_M SFGSDMMAWTLAMGHAMVRAGLTLSGPGAEQPWQPDIVHAHDWLVAHPAI
MUL_1919|M.ulcerans_Agy99 SFGSDMMAWTLAMGHAMVRAGLTLSGPGAEQPWQPDIVHAHDWLVAHPAI
Mvan_2096|M.vanbaalenii_PYR-1 TFGTDMMAWTLAMGHAMIRAGLVLAD------WRPDIVHAHDWLVATPAV
MAB_3366|M.abscessus_ATCC_1997 DFGTDMMAWTLAMGHAMVRAGLALGLKGSG-DWRPDVVHAHDWLVAHPAI
MAV_1024|M.avium_104 KVRSAITRIAKEDG--------------------VDIVHDHTFAGPMNAP
MSMEG_5641|M.smegmatis_MC2_155 KVRKAIERIAATEG--------------------VDIVHDHTFAGPLNAP
TH_0068|M.thermoresistible__bu RTRRAIETLAATDG--------------------LDVVHDHTLAGPLNAS
Mflv_0483|M.gilvum_PYR-GCK IKESAHHAFVELMSELADPASG------------VDVVHNHSLNPVP--V
. . *:** *
Mb3058|M.bovis_AF2122/97 ALAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVHAVESWLVRESDSLIT
Rv3032|M.tuberculosis_H37Rv ALAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVHAVESWLVRESDSLIT
MLBr_01715|M.leprae_Br4923 TLAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVHAVESWLVRESDSLIT
MMAR_1681|M.marinum_M ALADYYDVPMVSTIHATEAGRHSGWVSGDLSRQVHAVESWLVRESDSLIT
MUL_1919|M.ulcerans_Agy99 ALADYYDVPMVSTIHATEAGRHSGWVSGDLSRQVHAVESWLVRESDSLIT
Mvan_2096|M.vanbaalenii_PYR-1 ALAQFFDVPLVSTIHATEAGRHSGWVSGQVSRQVHALESWLVRDSDSLIT
MAB_3366|M.abscessus_ATCC_1997 ALAEHFDVPLVSTLHATEAGRHSGWVSGRISRQVHSVEWWLARESDSLIT
MAV_1024|M.avium_104 IYQG-LGVPTVVTVHG------------PIDDDLHPYYRELSGDVGLVAI
MSMEG_5641|M.smegmatis_MC2_155 FYRK-LGLPTVVTVHG------------PIDEDLYPYYRELGDDIGLIAI
TH_0068|M.thermoresistible__bu AYRG-LGLPVVLTTHG------------PVHEEMYDYYRTLGSDVHLVAI
Mflv_0483|M.gilvum_PYR-GCK LVARRLPVPVLTTLHT------------PPFPALEAAVGAVARSGQAFAA
:* : * * : : . .
Mb3058|M.bovis_AF2122/97 CSASMNDEITELFGPGLAEITVIRNGIDAARWPFAARRPRTG-PAELLYV
Rv3032|M.tuberculosis_H37Rv CSASMNDEITELFGPGLAEITVIRNGIDAARWPFAARRPRTG-PAELLYV
MLBr_01715|M.leprae_Br4923 CSASMCNEIIELFGPGLAEITVIRNGIDPARWPFAARRARTG-PAELLYV
MMAR_1681|M.marinum_M CSGSMSDEITELFGPGLAETAVIRNGIDAARWPFAVRKPRTG-PPELLYV
MUL_1919|M.ulcerans_Agy99 CSGSMSDEITELFGPGLAETAVIRNGIDVARWPFAVRKPRTG-PPELLYV
Mvan_2096|M.vanbaalenii_PYR-1 CSASMRDEITTLFGPELADITVIPNGIDTDGWPFAPHRPRPG-PAELLYF
MAB_3366|M.abscessus_ATCC_1997 CSASMLDEVTQLFGPELPQVHVIRNGIDVTRWVFAPRPPADGTPPVLLFV
MAV_1024|M.avium_104 S------DRQRQLAPELNWVGRVHNALRIDEWPFETEKHDYA-----LFL
MSMEG_5641|M.smegmatis_MC2_155 S------DRQRELAPDLNWVGRVHNALRIDDWPFNTVKKDYA-----LFL
TH_0068|M.thermoresistible__bu S------DRQRSLAPDLNWLGRVHNALRTDEWPFHPEKQDYA-----LFL
Mflv_0483|M.gilvum_PYR-GCK VSR---QAAAAWEHVGIRRMALVRNGIDTSRWPAGPGGEDLV------WS
: : *.: * :
Mb3058|M.bovis_AF2122/97 GRLEYEKGVHDAIAALPRLRRTHPGTTLTIAGEG--TQQDWLIDQARKHR
Rv3032|M.tuberculosis_H37Rv GRLEYEKGVHDAIAALPRLRRTHPGTTLTIAGEG--TQQDWLIDQARKHR
MLBr_01715|M.leprae_Br4923 GRLEYEKGVHDVIAALPRIRRSYPGTTLTIAGEG--TQQDWLVDQARKYK
MMAR_1681|M.marinum_M GRLEYEKGVHNAIAALPRIRRAHPGTTLTIAGEG--TQQDWLVEQARKHR
MUL_1919|M.ulcerans_Agy99 GRLEYEKGVHNAIAALPRIRRAHPGTTLTIAGEG--TQQDWLVEQARKHR
Mvan_2096|M.vanbaalenii_PYR-1 GRLEYEKGVHDLIAALPRIRRAHPGTTLTIAGDG--TQLDFLVEQARKHK
MAB_3366|M.abscessus_ATCC_1997 GRLEYEKGIHDAIAALPKIRRAHPGTVLAVAGDG--TQQDWLLEQARKYK
MAV_1024|M.avium_104 GRYAPYKGAHLALRAA-----HEAGIPLVLAGKCSEPSEKEYFEDCVRPL
MSMEG_5641|M.smegmatis_MC2_155 GRYAPYKGAHLALEAG-----HAAGMPLVLAGKCSEPPEIAYFDEQVKPL
TH_0068|M.thermoresistible__bu GRFTAAKGPDLALHAA-----HRAGIPLVLAGKCTEPAEKAYFRETVQPL
Mflv_0483|M.gilvum_PYR-GCK GRLLEEKGPHLAVDAA-----LRAGRRIVLAGPI---VDDHFFRRRIAPT
** ** . : * .* :.:** .
Mb3058|M.bovis_AF2122/97 VLRATRFVGHLDHTELLALLHRADAAVLPSHYEP-FGLVALEAAAAGTPL
Rv3032|M.tuberculosis_H37Rv VLRATRFVGHLDHTELLALLHRADAAVLPSHYEP-FGLVALEAAAAGTPL
MLBr_01715|M.leprae_Br4923 VIKATRFVGHLNHNELLAALQRADAAVLPSHYEP-FGLVALEAAAAGTPL
MMAR_1681|M.marinum_M VLKATKFLGHLEHAELLTLLHRADAAVFPSHYEP-FGLVALEAAAAGTPL
MUL_1919|M.ulcerans_Agy99 VLKATKFLGHLEHAELLTLLHRADAAVFPSHYEP-FGLVALEAAAAGTPL
Mvan_2096|M.vanbaalenii_PYR-1 VVKATNFIGRVGHSELLRLLHRADAAVLPSHYEP-FGIVALEAAATGTPL
MAB_3366|M.abscessus_ATCC_1997 VVKSVQFLGTLDHSELLHWLHHADAILLPSHYEP-FGIVALEAAAAGTPL
MAV_1024|M.avium_104 LTDNDHVFGEADAVSKRKLLSRARCLLFPIQWEEPFGIVMIEAMVCGTPV
MSMEG_5641|M.smegmatis_MC2_155 LTDSDHVFGMADAQAKRKLLAEARCLVFPIQWEEPFGMVMIEAMACGTPV
TH_0068|M.thermoresistible__bu LTDADLLFGQADARAKRRLLSRARCLVFPVQWEEPFGMVMIESMVCGTPV
Mflv_0483|M.gilvum_PYR-GCK LGSRASYAGHLGQRELATLVGRAAAALVTPLWDEPYGQVVVEAMACGTPV
: * : .* . :.. :: :* * :*: . ***:
Mb3058|M.bovis_AF2122/97 VTSNIGGLGEAVINGQTGVSCAPRDVAGLAAAVRSVLDDPAAAQRRARAA
Rv3032|M.tuberculosis_H37Rv VTSNIGGLGEAVINGQTGVSCAPRDVAGLAAAVRSVLDDPAAAQRRARAA
MLBr_01715|M.leprae_Br4923 VTSNIGGLGEAVINGQTGVSCPPRDIAELAAMVCTVLEDPDAAQQRALAA
MMAR_1681|M.marinum_M VTSNIGGLGEAVIDGVTGVSCPPRDVAALAAAVCGVLDDPAAAQQRARAA
MUL_1919|M.ulcerans_Agy99 VTSNIGGLGEAVIDGVTGVSCPPRDVAALAAAVCGVLDDPAAAQQRARAA
Mvan_2096|M.vanbaalenii_PYR-1 VTSTAGGLGEAVIDGVTGLSYPPGDVAALARAVRAVLDSPAAAQRRARAA
MAB_3366|M.abscessus_ATCC_1997 VTSTVGGLGEAVIDGETGMSFAPRDVAGIAEAVTRTLDDPAAAAERAVAA
MAV_1024|M.avium_104 VALRGGAVSEVVADGVTGVVCDHPDELPGAIARAQTLD-PHACRRHVAAN
MSMEG_5641|M.smegmatis_MC2_155 VALRGGAVPEVIVDGVTGVICDVPEELPAAMEKATHLD-PAACRRHVAAH
TH_0068|M.thermoresistible__bu VALRGGAVSEVVVDGVTGFVCDDAADLADAIKEASRLD-PYACRRHVEAN
Mflv_0483|M.gilvum_PYR-GCK VAFAGGGIPELVDEHS-GRLVEPGDVAALAAAVREAVALPRAGVRRVAVQ
*: *.: * : : * * : * * .:. .
Mb3058|M.bovis_AF2122/97 -RQRLTSDFDWQTVATATAQVYLAAKRGERQP--QPRLPIVEHALPDR--
Rv3032|M.tuberculosis_H37Rv -RQRLTSDFDWQTVATATAQVYLAAKRGERQP--QPRLPIVEHALPDR--
MLBr_01715|M.leprae_Br4923 -RERLTSDFDWQTVAQQTAQVYLAAKRRERQP--QPRLPIVEHALPDR--
MMAR_1681|M.marinum_M -RERLNSDFDWQTVAEQTAQVYLAAKRGERQP--LPRLPIVEHALPDR--
MUL_1919|M.ulcerans_Agy99 -RERLNSNFDWQTVAEQTAQVYLAAKRGERQP--LPRLPIVEHALPDR--
Mvan_2096|M.vanbaalenii_PYR-1 -RERLTSDFSWHTVAAETAQVYLAAKRAVRRP--QPRRPIEERPLPDR--
MAB_3366|M.abscessus_ATCC_1997 -RARLTADFDWATVADETAQVYLAAKRREREP--LGRSVIVERPLPDR--
MAV_1024|M.avium_104 -FAVAQFGSAYEAIYRNVLQHATAAPRRMRSTGLVESIETGRRACDFRQK
MSMEG_5641|M.smegmatis_MC2_155 -FGVGRFGAGYEQIYRNVLRQRTRMTLREVLT---EPPGVAERASA----
TH_0068|M.thermoresistible__bu -FGVDQLAVGYEQVYRQAMAAMATPGS--------DRPTLGMSA------
Mflv_0483|M.gilvum_PYR-GCK RHGLARMVTGYVALYRALIAEATPPPRPPKLHSSSESANQSQAVSATARA
: :
Mb3058|M.bovis_AF2122/97 ------------
Rv3032|M.tuberculosis_H37Rv ------------
MLBr_01715|M.leprae_Br4923 ------------
MMAR_1681|M.marinum_M ------------
MUL_1919|M.ulcerans_Agy99 ------------
Mvan_2096|M.vanbaalenii_PYR-1 ------------
MAB_3366|M.abscessus_ATCC_1997 ------------
MAV_1024|M.avium_104 APA---------
MSMEG_5641|M.smegmatis_MC2_155 ------------
TH_0068|M.thermoresistible__bu ------------
Mflv_0483|M.gilvum_PYR-GCK CLTNVSGVSAET