For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KILMVSWEYPPVVIGGLGRHVHHLATALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVIAAAQDPH EFSFGSDMMAWTLAMGHAMVRAGLTLSGPGAEQPWQPDIVHAHDWLVAHPAIALADYYDVPMVSTIHATE AGRHSGWVSGDLSRQVHAVESWLVRESDSLITCSGSMSDEITELFGPGLAETAVIRNGIDAARWPFAVRK PRTGPPELLYVGRLEYEKGVHNAIAALPRIRRAHPGTTLTIAGEGTQQDWLVEQARKHRVLKATKFLGHL EHAELLTLLHRADAAVFPSHYEPFGLVALEAAAAGTPLVTSNIGGLGEAVIDGVTGVSCPPRDVAALAAA VCGVLDDPAAAQQRARAARERLNSDFDWQTVAEQTAQVYLAAKRGERQPLPRLPIVEHALPDR*
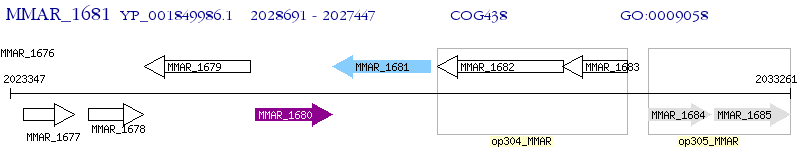
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1681 | - | - | 100% (414) | transferase |
| M. marinum M | MMAR_0812 | - | 1e-29 | 29.52% (393) | mannosyltransferase |
| M. marinum M | MMAR_4226 | - | 7e-29 | 28.40% (419) | glycosyl transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3058 | - | 0.0 | 86.71% (414) | transferase |
| M. gilvum PYR-GCK | Mflv_4262 | - | 0.0 | 76.33% (414) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv3032 | - | 0.0 | 86.71% (414) | transferase |
| M. leprae Br4923 | MLBr_01715 | - | 0.0 | 84.06% (414) | putative transferase |
| M. abscessus ATCC 19977 | MAB_3366 | - | 0.0 | 74.70% (415) | glycosyltransferase |
| M. avium 104 | MAV_3879 | - | 0.0 | 83.73% (418) | glycosyl transferase, group 1 family protein |
| M. smegmatis MC2 155 | MSMEG_0933 | - | 5e-30 | 28.89% (405) | hypothetical protein MSMEG_0933 |
| M. thermoresistible (build 8) | TH_2191 | - | 0.0 | 79.57% (416) | POSSIBLE TRANSFERASE |
| M. ulcerans Agy99 | MUL_1919 | - | 0.0 | 99.52% (414) | transferase |
| M. vanbaalenii PYR-1 | Mvan_2096 | - | 0.0 | 77.24% (413) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4262|M.gilvum_PYR-GCK ----------------------MKMKILLVSWEYPPVVIG------GLGR
Mvan_2096|M.vanbaalenii_PYR-1 ----------------------MKTKILLVSWEYPPVVIG------GLGR
MMAR_1681|M.marinum_M ------------------------MKILMVSWEYPPVVIG------GLGR
MUL_1919|M.ulcerans_Agy99 ------------------------MKILMVSWEYPPVVIG------GLGR
Mb3058|M.bovis_AF2122/97 ------------------------MRILMVSWEYPPVVIG------GLGR
Rv3032|M.tuberculosis_H37Rv ------------------------MRILMVSWEYPPVVIG------GLGR
MLBr_01715|M.leprae_Br4923 MFIQRHAVVRGIFGTFAPAEGTTLMKILIVSWEYPPVVIG------GLGR
MAV_3879|M.avium_104 ------------------------MKILMVSWEYPPVVIG------GLGR
TH_2191|M.thermoresistible__bu ------------------------MKILMVSWEYPPVVVG------GLGR
MAB_3366|M.abscessus_ATCC_1997 ------------------------MRILMVSWEYPPVVIG------GLGR
MSMEG_0933|M.smegmatis_MC2_155 --------------MRLATDLETPRRVAVLSVHTSPLAQPGTGDAGGMNV
:: ::* . .*:. *:.
Mflv_4262|M.gilvum_PYR-GCK HVHHLATELVAAGHEVVVLSRRPTGTDPQTHPTTDETHEGVRVIAAAEDP
Mvan_2096|M.vanbaalenii_PYR-1 HVHHLATELAQSGHEVVVLSRRPTATDPQTHPTTDEVHEGVRVIAAAEDP
MMAR_1681|M.marinum_M HVHHLATALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVIAAAQDP
MUL_1919|M.ulcerans_Agy99 HVHHLATALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVIAAAQDP
Mb3058|M.bovis_AF2122/97 HVHHLSTALAAAGHDVVVLSRCPSGTDPSTHPSSDEVTEGVRVIAAAQDP
Rv3032|M.tuberculosis_H37Rv HVHHLSTALAAAGHDVVVLSRCPSGTDPSTHPSSDEVTEGVRVIAAAQDP
MLBr_01715|M.leprae_Br4923 HVHHLSTALAAAGHDVVVLSRRPSGTDPCTHPTSDEISEGVRVIAAAQDP
MAV_3879|M.avium_104 HVHHLSTALAAAGHDVVVLSRRPTGTDPRTHPSTDQISEGVRVIAAAQDP
TH_2191|M.thermoresistible__bu HVYHLSTALAAAGHEVVVLSRRPTGTDPSTHPSTDEVRDGVRVLAAAQDP
MAB_3366|M.abscessus_ATCC_1997 HVHHLSTELAAAGHDVVVLTRRPSGTDPSSHPTSDEISEGVRVVAAAEDP
MSMEG_0933|M.smegmatis_MC2_155 YVLQTALQLARRGVEVEVFTRATSSADAPVVP----VAPGVLVRNVVAGP
:* : : *. * :* *::* .:.:*. * ** * .. .*
Mflv_4262|M.gilvum_PYR-GCK HEFGFGTDMMAWVLAMGHSMIRAGFG----------LDGWRPDIVHAHDW
Mvan_2096|M.vanbaalenii_PYR-1 HEFTFGTDMMAWTLAMGHAMIRAGLV----------LADWRPDIVHAHDW
MMAR_1681|M.marinum_M HEFSFGSDMMAWTLAMGHAMVRAGLT----LSGPGAEQPWQPDIVHAHDW
MUL_1919|M.ulcerans_Agy99 HEFSFGSDMMAWTLAMGHAMVRAGLT----LSGPGAEQPWQPDIVHAHDW
Mb3058|M.bovis_AF2122/97 HEFTFGNDMMAWTLAMGHAMIRAGLR----LKKLGTDRSWRPDVVHAHDW
Rv3032|M.tuberculosis_H37Rv HEFTFGNDMMAWTLAMGHAMIRAGLR----LKKLGTDRSWRPDVVHAHDW
MLBr_01715|M.leprae_Br4923 HEFTFSNDMMAWTLAMGHAMIRTGLS----LTRHSSDLPWRPDVVHAHDW
MAV_3879|M.avium_104 HEFTFGTDMMAWTLAMGHAMIRAGLPPKLHDKKPGGNRPWRPDVVHAHDW
TH_2191|M.thermoresistible__bu HEFDFGRDMMAWTLAMGHSMVRAGLALG--LRGGTGGTPWQPDVVHAHDW
MAB_3366|M.abscessus_ATCC_1997 HDFDFGTDMMAWTLAMGHAMVRAGLALG-----LKGSGDWRPDVVHAHDW
MSMEG_0933|M.smegmatis_MC2_155 FEGLDKNDLPTQLCAFTAGVLRAEAT----------HEPGYYDVVHSHYW
.: *: : *: .::*: *:**:* *
Mflv_4262|M.gilvum_PYR-GCK LVAHPAVALAQFFDVPLVSTIHATEAGRHSGWVSGSVSR--QVHALESWL
Mvan_2096|M.vanbaalenii_PYR-1 LVATPAVALAQFFDVPLVSTIHATEAGRHSGWVSGQVSR--QVHALESWL
MMAR_1681|M.marinum_M LVAHPAIALADYYDVPMVSTIHATEAGRHSGWVSGDLSR--QVHAVESWL
MUL_1919|M.ulcerans_Agy99 LVAHPAIALADYYDVPMVSTIHATEAGRHSGWVSGDLSR--QVHAVESWL
Mb3058|M.bovis_AF2122/97 LVAHPAIALAQFYDVPMVSTIHATEAGRHSGWVSGALSR--QVHAVESWL
Rv3032|M.tuberculosis_H37Rv LVAHPAIALAQFYDVPMVSTIHATEAGRHSGWVSGALSR--QVHAVESWL
MLBr_01715|M.leprae_Br4923 LVAHPAITLAQFYDVPMVSTIHATEAGRHSGWVSGALSR--QVHAVESWL
MAV_3879|M.avium_104 LVAHPAIALAQFYDVPMVSTIHATEAGRHSGWVSGPISR--QVHAIESWL
TH_2191|M.thermoresistible__bu LVAHPAIALAEFFDVPLVSTIHATEAGRHSGWVSGPISR--QVHAVESWL
MAB_3366|M.abscessus_ATCC_1997 LVAHPAIALAEHFDVPLVSTLHATEAGRHSGWVSGRISR--QVHSVEWWL
MSMEG_0933|M.smegmatis_MC2_155 LSGQVGWLARDRWAVPLVHTAHTLAAVKNAALAAGDAPEPPLRAVGEQQV
* . . : : **:* * *: * :::. .:* .. * :
Mflv_4262|M.gilvum_PYR-GCK VRDSDSLITCSASMRDEISALFGPELAEISVIPNGIDTDGWPFAARRAHS
Mvan_2096|M.vanbaalenii_PYR-1 VRDSDSLITCSASMRDEITTLFGPELADITVIPNGIDTDGWPFAPHRPRP
MMAR_1681|M.marinum_M VRESDSLITCSGSMSDEITELFGPGLAETAVIRNGIDAARWPFAVRKPRT
MUL_1919|M.ulcerans_Agy99 VRESDSLITCSGSMSDEITELFGPGLAETAVIRNGIDVARWPFAVRKPRT
Mb3058|M.bovis_AF2122/97 VRESDSLITCSASMNDEITELFGPGLAEITVIRNGIDAARWPFAARRPRT
Rv3032|M.tuberculosis_H37Rv VRESDSLITCSASMNDEITELFGPGLAEITVIRNGIDAARWPFAARRPRT
MLBr_01715|M.leprae_Br4923 VRESDSLITCSASMCNEIIELFGPGLAEITVIRNGIDPARWPFAARRART
MAV_3879|M.avium_104 VRESDSLITCSASMADEITELFGPGLAQITVIRNGIETARWPFAPRRPRT
TH_2191|M.thermoresistible__bu VRDSDSLITCSASMSEEITELFGPGLSEIRVIRNGIDVAQWPFARRSPRS
MAB_3366|M.abscessus_ATCC_1997 ARESDSLITCSASMLDEVTQLFGPELPQVHVIRNGIDVTRWVFAPRPPAD
MSMEG_0933|M.smegmatis_MC2_155 VDEADRLIVNTEVEAQQLVSLHNADRSRIDVVHPGVDLDVFTPGSRDAAR
. ::* **. : ::: *... . *: *:: : . : .
Mflv_4262|M.gilvum_PYR-GCK G-------PAELLYFGRLEYEKGVHDAIAALPKVRRTHPGTTLTIAGDG-
Mvan_2096|M.vanbaalenii_PYR-1 G-------PAELLYFGRLEYEKGVHDLIAALPRIRRAHPGTTLTIAGDG-
MMAR_1681|M.marinum_M G-------PPELLYVGRLEYEKGVHNAIAALPRIRRAHPGTTLTIAGEG-
MUL_1919|M.ulcerans_Agy99 G-------PPELLYVGRLEYEKGVHNAIAALPRIRRAHPGTTLTIAGEG-
Mb3058|M.bovis_AF2122/97 G-------PAELLYVGRLEYEKGVHDAIAALPRLRRTHPGTTLTIAGEG-
Rv3032|M.tuberculosis_H37Rv G-------PAELLYVGRLEYEKGVHDAIAALPRLRRTHPGTTLTIAGEG-
MLBr_01715|M.leprae_Br4923 G-------PAELLYVGRLEYEKGVHDVIAALPRIRRSYPGTTLTIAGEG-
MAV_3879|M.avium_104 G-------PAELLYLGRLEYEKGVHDVIAALPRIRRTHPGTTLTIAGEG-
TH_2191|M.thermoresistible__bu G-------PPRLLYVGRLEYEKGVQDAIAALPRIRRTHPGTTLTIAGTG-
MAB_3366|M.abscessus_ATCC_1997 GT------PPVLLFVGRLEYEKGIHDAIAALPKIRRAHPGTVLAVAGDG-
MSMEG_0933|M.smegmatis_MC2_155 AVFGLPTDQKIVAFVGRIQPLKAPDILLRAAAKLPGVRVLIAGGPSGSGL
. : :.**:: *. . : * .:: . :* *
Mflv_4262|M.gilvum_PYR-GCK TQLGFLTEQARRHKVLKATKFVGRVDHSQLLTLLHRADVAVLPSHYEPFG
Mvan_2096|M.vanbaalenii_PYR-1 TQLDFLVEQARKHKVVKATNFIGRVGHSELLRLLHRADAAVLPSHYEPFG
MMAR_1681|M.marinum_M TQQDWLVEQARKHRVLKATKFLGHLEHAELLTLLHRADAAVFPSHYEPFG
MUL_1919|M.ulcerans_Agy99 TQQDWLVEQARKHRVLKATKFLGHLEHAELLTLLHRADAAVFPSHYEPFG
Mb3058|M.bovis_AF2122/97 TQQDWLIDQARKHRVLRATRFVGHLDHTELLALLHRADAAVLPSHYEPFG
Rv3032|M.tuberculosis_H37Rv TQQDWLIDQARKHRVLRATRFVGHLDHTELLALLHRADAAVLPSHYEPFG
MLBr_01715|M.leprae_Br4923 TQQDWLVDQARKYKVIKATRFVGHLNHNELLAALQRADAAVLPSHYEPFG
MAV_3879|M.avium_104 TQQGWLIEQARKHRVLKAIRFVGHLEHSELLAMLHRADAAVLPSHYEPFG
TH_2191|M.thermoresistible__bu TQHEWLVEQARKHKVLKATRFVGQLGHEALLRQLHTADAAVLPSHYEPFG
MAB_3366|M.abscessus_ATCC_1997 TQQDWLLEQARKYKVVKSVQFLGTLDHSELLHWLHHADAILLPSHYEPFG
MSMEG_0933|M.smegmatis_MC2_155 AQPDTLVRLADELGISDRVTFLPPQSREQLVNVYRAADLVAVPSYSESFG
:* * * . : *: : *: : ** .**: *.**
Mflv_4262|M.gilvum_PYR-GCK IVALEAAATGTPLVTSTAGGLGEAVIDGVTGMSYPPSDVIALAAAIRAVL
Mvan_2096|M.vanbaalenii_PYR-1 IVALEAAATGTPLVTSTAGGLGEAVIDGVTGLSYPPGDVAALARAVRAVL
MMAR_1681|M.marinum_M LVALEAAAAGTPLVTSNIGGLGEAVIDGVTGVSCPPRDVAALAAAVCGVL
MUL_1919|M.ulcerans_Agy99 LVALEAAAAGTPLVTSNIGGLGEAVIDGVTGVSCPPRDVAALAAAVCGVL
Mb3058|M.bovis_AF2122/97 LVALEAAAAGTPLVTSNIGGLGEAVINGQTGVSCAPRDVAGLAAAVRSVL
Rv3032|M.tuberculosis_H37Rv LVALEAAAAGTPLVTSNIGGLGEAVINGQTGVSCAPRDVAGLAAAVRSVL
MLBr_01715|M.leprae_Br4923 LVALEAAAAGTPLVTSNIGGLGEAVINGQTGVSCPPRDIAELAAMVCTVL
MAV_3879|M.avium_104 IVALEAAAAGTPLVTSNIGGLGEAVIDGQTGVSCPPRDVPALAAAIRTVL
TH_2191|M.thermoresistible__bu IVALEAAATGAPLVTSTAGGLGEAVIDGVTGKSFKPRDIAGLTAAVRAVL
MAB_3366|M.abscessus_ATCC_1997 IVALEAAAAGTPLVTSTVGGLGEAVIDGETGMSFAPRDVAGIAEAVTRTL
MSMEG_0933|M.smegmatis_MC2_155 LVAVEAQACGTPVVAAAVGGLPVAVADGVSGALVDGHDIGDWADTISEVL
:**:** * *:*:*:: *** ** :* :* *: : : .*
Mflv_4262|M.gilvum_PYR-GCK DAPAAAQERATAARARLTSDFAWHTVAAETAQVYLAAK--RAERRPQPRR
Mvan_2096|M.vanbaalenii_PYR-1 DSPAAAQRRARAARERLTSDFSWHTVAAETAQVYLAAK--RAVRRPQPRR
MMAR_1681|M.marinum_M DDPAAAQQRARAARERLNSDFDWQTVAEQTAQVYLAAK--RGERQPLPRL
MUL_1919|M.ulcerans_Agy99 DDPAAAQQRARAARERLNSNFDWQTVAEQTAQVYLAAK--RGERQPLPRL
Mb3058|M.bovis_AF2122/97 DDPAAAQRRARAARQRLTSDFDWQTVATATAQVYLAAK--RGERQPQPRL
Rv3032|M.tuberculosis_H37Rv DDPAAAQRRARAARQRLTSDFDWQTVATATAQVYLAAK--RGERQPQPRL
MLBr_01715|M.leprae_Br4923 EDPDAAQQRALAARERLTSDFDWQTVAQQTAQVYLAAK--RRERQPQPRL
MAV_3879|M.avium_104 DDPEAAQRRACAARERLTSDFDWHTVADKTAQVYLSAK--RAERQPQARL
TH_2191|M.thermoresistible__bu DDPEGAQQRAVAARERLTADFDWHSVAAETAQVYLAAK--RREREPLPRR
MAB_3366|M.abscessus_ATCC_1997 DDPAAAAERAVAARARLTADFDWATVADETAQVYLAAK--RREREPLGRS
MSMEG_0933|M.smegmatis_MC2_155 DREPAALSRASAEHA---AQFSWAHTVDALLASYSRAMSDYRARHPRPAA
: .* ** * : ::* * .. * * *.*
Mflv_4262|M.gilvum_PYR-GCK TIVERPLPDR-----
Mvan_2096|M.vanbaalenii_PYR-1 PIEERPLPDR-----
MMAR_1681|M.marinum_M PIVEHALPDR-----
MUL_1919|M.ulcerans_Agy99 PIVEHALPDR-----
Mb3058|M.bovis_AF2122/97 PIVEHALPDR-----
Rv3032|M.tuberculosis_H37Rv PIVEHALPDR-----
MLBr_01715|M.leprae_Br4923 PIVEHALPDR-----
MAV_3879|M.avium_104 PIVEHALPDR-----
TH_2191|M.thermoresistible__bu LIVEHALPDR-----
MAB_3366|M.abscessus_ATCC_1997 VIVERPLPDR-----
MSMEG_0933|M.smegmatis_MC2_155 RRSGRRFSMRRGVRT
: :. *