For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGNDILAPADVDDPLRIVLVAPPYFDIPPKGYGGTEAVVADLADTLTARGHHVTVLGAGAPGTAARFQPL WERTLPDRLGQPYPEIMHALKVRKAIERIAATEGVDIVHDHTFAGPLNAPFYRKLGLPTVVTVHGPIDED LYPYYRELGDDIGLIAISDRQRELAPDLNWVGRVHNALRIDDWPFNTVKKDYALFLGRYAPYKGAHLALE AGHAAGMPLVLAGKCSEPPEIAYFDEQVKPLLTDSDHVFGMADAQAKRKLLAEARCLVFPIQWEEPFGMV MIEAMACGTPVVALRGGAVPEVIVDGVTGVICDVPEELPAAMEKATHLDPAACRRHVAAHFGVGRFGAGY EQIYRNVLRQRTRMTLREVLTEPPGVAERASA*
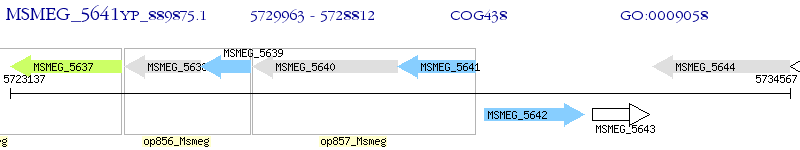
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5641 | - | - | 100% (383) | glycosyl transferase, group 1 family protein |
| M. smegmatis MC2 155 | MSMEG_2935 | - | 4e-12 | 26.49% (336) | phosphatidylinositol alpha-mannosyltransferase |
| M. smegmatis MC2 155 | MSMEG_5080 | - | 5e-12 | 27.67% (347) | glycogen synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1244c | - | 9e-15 | 28.76% (379) | putative glycosyl transferase |
| M. gilvum PYR-GCK | Mflv_0483 | - | 3e-32 | 30.19% (361) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv1212c | - | 7e-15 | 29.02% (379) | putative glycosyl transferase |
| M. leprae Br4923 | MLBr_00452 | - | 2e-14 | 26.92% (338) | putative glycosyltransferase |
| M. abscessus ATCC 19977 | MAB_2894c | - | 8e-13 | 27.49% (342) | phosphatidylinositol alpha-mannosyltransferase |
| M. marinum M | MMAR_4226 | - | 3e-16 | 30.16% (378) | glycosyl transferase |
| M. avium 104 | MAV_1024 | - | 1e-167 | 75.82% (364) | glycosyl transferase, group 1 family protein |
| M. thermoresistible (build 8) | TH_0092 | - | 1e-167 | 75.60% (373) | glycosyl transferase, group 1 family protein |
| M. ulcerans Agy99 | MUL_3252 | pimA | 1e-14 | 26.76% (355) | alpha-mannosyltransferase PimA |
| M. vanbaalenii PYR-1 | Mvan_2096 | - | 1e-12 | 26.09% (414) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5641|M.smegmatis_MC2_155 MSGN------------------------DILAPADVDD---PLRIVLVAP
TH_0092|M.thermoresistible__bu MSGNGFRFHHTFRDQNATAADTAPADTAALLSETESTD---PLRIVLVAP
MAV_1024|M.avium_104 MRGA------------------------DFVLPTDVPEAGRPLRIVLVAP
MLBr_00452|M.leprae_Br4923 ------------------------------------------MRIGMICP
MUL_3252|M.ulcerans_Agy99 ------------------MAHVSATMVGRLVRCPPRAAAGALMRIGMVCP
MAB_2894c|M.abscessus_ATCC_199 ------------------------------------------MRIGMVCP
Mb1244c|M.bovis_AF2122/97 ------------------------------------------MRVAMLTR
Rv1212c|M.tuberculosis_H37Rv ------------------------------------------MRVAMLTR
MMAR_4226|M.marinum_M ------------------------------------------MRVAMLTR
Mflv_0483|M.gilvum_PYR-GCK ------------------------------------------MRVALIAP
Mvan_2096|M.vanbaalenii_PYR-1 ------------------------------------------MKTKILLV
:: ::
MSMEG_5641|M.smegmatis_MC2_155 PYFDIPPKGYGGTEAVVADLADTLTARGHHVTVLGA------GAP-GTAA
TH_0092|M.thermoresistible__bu PYFDIPPKAYGGTEAVVADLADSLTARGHRVTVLGA------GNP-RIAA
MAV_1024|M.avium_104 PYFDVPPDGYGGVEAVVADLADMLVARGHQVTLLGA------GEP-GTAA
MLBr_00452|M.leprae_Br4923 YSFDVP----GGVQSHVLQLAEVMRARGQQVRVLAP------ASPDVSLP
MUL_3252|M.ulcerans_Agy99 YSFDVA----GGVQSHVLQLAEVMRARGHEVSVLAP------ASPHAALP
MAB_2894c|M.abscessus_ATCC_199 YSFDVP----GGVQSHVVQLAEVMRDRGHHVSLLAP------SSSDLELP
Mb1244c|M.bovis_AF2122/97 ---EYPPEVYGGAGVHVTELVAYLR-RLCAVDVHCM------GAPRPGAF
Rv1212c|M.tuberculosis_H37Rv ---EYPPEVYGGAGVHVTELVAYLR-RLCAVDVHCM------GAPRPGAF
MMAR_4226|M.marinum_M ---EYPPEVYGGAGVHVTELVARLQ-RLCTVDVHCM------GAPRPDAS
Mflv_0483|M.gilvum_PYR-GCK HHFPIREPFAGGIESHVWHLARALIRQGHAVTLCAA------AGSDLAME
Mvan_2096|M.vanbaalenii_PYR-1 S-WEYPPVVIGGLGRHVHHLATELAQSGHEVVVLSRRPTATDPQTHPTTD
** * .*. : * : .
MSMEG_5641|M.smegmatis_MC2_155 R------FQPLWERTLPDRLGQPYPEIMHALKVRKAIERIAATEG-VDIV
TH_0092|M.thermoresistible__bu R------FVPLWERTIPDRLGQPYPEVMHALKVRRAIADIADREGGIDLI
MAV_1024|M.avium_104 R------FIPLWDRTVPERLGEPYPEIMHALKVRSAITRIAKEDG-VDIV
MLBr_00452|M.leprae_Br4923 E------YVVSAGRAIPIPYNGSVARLQFSPAVHSRVRRWLVDGD-FDVL
MUL_3252|M.ulcerans_Agy99 D------YVVSGGKAVPIPYNGSVARLRFGPATHRKVKKWLSEGD-FDVL
MAB_2894c|M.abscessus_ATCC_199 E------FVVSGGKAVPIPYNGSVARLRFGPATYAKTRRWLVDGD-FDVL
Mb1244c|M.bovis_AF2122/97 A------YRP----------DPRLGSANAALSTLSADLVMANAASAATVV
Rv1212c|M.tuberculosis_H37Rv A------YRP----------DPRLGSANAALSTLSADLVMANAASAATVV
MMAR_4226|M.marinum_M A------HQP----------DPRLANANPALATLSADLVIANAADAASVV
Mflv_0483|M.gilvum_PYR-GCK HPALTVTAMPRSAAASRVFPPPGAIKESAHHAFVELMSELADPASGVDVV
Mvan_2096|M.vanbaalenii_PYR-1 EVHEGVRVIAAAEDPHEFTFGTDMMAWTLAMGHAMIRAGLVLADWRPDIV
::
MSMEG_5641|M.smegmatis_MC2_155 HDHTFAGP-LNAPFYRKLGLPTVVTVHG--------------PIDEDLYP
TH_0092|M.thermoresistible__bu HDHTFAGP-LNVPAYRELGVPTVATVHG--------------PIDEDLYP
MAV_1024|M.avium_104 HDHTFAGP-MNAPIYQGLGVPTVVTVHG--------------PIDDDLHP
MLBr_00452|M.leprae_Br4923 HLHEPNAPSLSMWALRVAEGPIVATFHTST-----TKSLTLSVFQGVLRP
MUL_3252|M.ulcerans_Agy99 HLHEPNAPSLSMLALNIAEGPIVATFHTST-----TKSLTLTVFQSILRP
MAB_2894c|M.abscessus_ATCC_199 HLHEPNAPSLSMLALMVAEGPIVATFHTST-----TKSLTLSVFQAVLRP
Mb1244c|M.bovis_AF2122/97 HSHTWYTALAGHLAAILYDIPHILTAHSLEPLRPWKKEQLGGGYQVSTWV
Rv1212c|M.tuberculosis_H37Rv HSHTWYTALAGHLAAILYDIPHVLTAHSLEPLRPWKKEQLGGGYQVSTWV
MMAR_4226|M.marinum_M HSHTWYTGLAGHLAALLYGIPHVLTAHSLEPMRPWKTEQLGGGYQISSWV
Mflv_0483|M.gilvum_PYR-GCK HNHSLNPV--PVLVARRLPVPVLTTLHTP-------------PFPALEAA
Mvan_2096|M.vanbaalenii_PYR-1 HAHDWLVATPAVALAQFFDVPLVSTIHATEAG--RHSGWVSGQVSRQVHA
* * * : * *
MSMEG_5641|M.smegmatis_MC2_155 YYRELGDDIGLIAISDRQR------ELAPDLNWVGRVHNALRIDDWPFN-
TH_0092|M.thermoresistible__bu YYRELGHDVGLIAISDRQR------ELAPDLNWIGRVHNALRIHDWPFK-
MAV_1024|M.avium_104 YYRELSGDVGLVAISDRQR------QLAPELNWVGRVHNALRIDEWPFE-
MLBr_00452|M.leprae_Br4923 WHEKIIGRIAVSDLARRWQ------MEALGSDAV-EIPNGVNVDSLSSA-
MUL_3252|M.ulcerans_Agy99 MHEKIVGRIAVSDLARRWQ------MEALGSDAV-EIPNGVDVGSFASS-
MAB_2894c|M.abscessus_ATCC_199 WHEKIVGRIAVSELARRWQ------MEALGSDAV-EIPNGVDVRSFSGA-
Mb1244c|M.bovis_AF2122/97 EQTAVLAANAVIAVSSAMRNDMLRVYPSLDPNLVHVIRNGIDTETWYPAG
Rv1212c|M.tuberculosis_H37Rv EQTAVLAANAVIAVSSAMRNDMLRVYPSLDPNLVHVIRNGIDTETWYPAG
MMAR_4226|M.marinum_M EKTAVLAADAVIAVSSGMRDDVLRLYPGLDPGMVHVVRNGIDTEVWFPAG
Mflv_0483|M.gilvum_PYR-GCK VGAVARSGQAFAAVSRQAAA----AWEHVGIRRMALVRNGIDTSRWPAG-
Mvan_2096|M.vanbaalenii_PYR-1 LESWLVRDSDSLITCSASMRDEITTLFGPELADITVIPNGIDTDGWPFAP
. : : *.:
MSMEG_5641|M.smegmatis_MC2_155 -------TVKKDY------ALFLGRY-APYKGAHLALEAGH-AAGMPLVL
TH_0092|M.thermoresistible__bu -------REKGDY------ALFLGRF-APYKGAHLAIQAAD-RAGIRLVL
MAV_1024|M.avium_104 -------TEKHDY------ALFLGRY-APYKGAHLALRAAH-EAGIPLVL
MLBr_00452|M.leprae_Br4923 -------PQLAGYPRLGKTVLFLGRYDEPRKGMSVLLDALP-GVMECFDD
MUL_3252|M.ulcerans_Agy99 -------SCLDGYPRPGKTVLFLGRYDEPRKGMSVLLEALP-KLVERFID
MAB_2894c|M.abscessus_ATCC_199 -------PVLEGYPREGKTVLFLGRYDEPRKGMDVLMGALP-KIATSFPD
Mb1244c|M.bovis_AF2122/97 PARTGSVLAELGVDPNRPMAVFVGRITRQKGVVHLVTAAHRFRSDVQLVL
Rv1212c|M.tuberculosis_H37Rv PARTGSVLAELGVDPNRPMAVFVGRITRQKGVVHLVTAAHRFRSDVQLVL
MMAR_4226|M.marinum_M PVGTASVLAELGVDPNRPIVAFVGRITRQKGVPHLLAAAHQFSPDVQLVL
Mflv_0483|M.gilvum_PYR-GCK --------------PGGEDLVWSGRLLEEKG-PHLAVDAAL-RAGRRIVL
Mvan_2096|M.vanbaalenii_PYR-1 ----------HRPRPGPAELLYFGRLEYEKGVHDLIAALPRIRRAHPGTT
: ** :
MSMEG_5641|M.smegmatis_MC2_155 AGKCSEPPEIAYFDEQVKPLLTDSDH----VFGMADAQAKRKLLAEARCL
TH_0092|M.thermoresistible__bu AGKCDEPSEKAYFEEQVRPLLNENDE----VFGEADAIAKRELLANARCL
MAV_1024|M.avium_104 AGKCSEPSEKEYFEDCVRPLLTDNDH----VFGEADAVSKRKLLSRARCL
MLBr_00452|M.leprae_Br4923 VQLLIVGRGDEEQLRSQAGGLVEHIR----FLGQVDDAGKAAAMRSADVY
MUL_3252|M.ulcerans_Agy99 VQLLIVGRGDEDELRDEAGELAEHMR----FLGQVDDAGKASAMRSADVY
MAB_2894c|M.abscessus_ATCC_199 VEILIVGRGDEAELRTQAGPLAKHLK----FLGQVDDATKASAMRSADVY
Mb1244c|M.bovis_AF2122/97 CAGAADTPEVADEVRVAVAELARNRTGVFWIQDRLTIGQLREILSAATVF
Rv1212c|M.tuberculosis_H37Rv CAGAADTPEVADEVRVAVAELARNRTGVFWIQDRLTIGQLREILSAATVF
MMAR_4226|M.marinum_M CAGAPDTPEIANEVQSAVAQLASTRSGVFWIRDILPVQKLREILSAATVF
Mflv_0483|M.gilvum_PYR-GCK AGPIVD---DHFFRRRIAPTLGSRAS----YAGHLGQRELATLVGRAAAA
Mvan_2096|M.vanbaalenii_PYR-1 LTIAGDGTQLDFLVEQARKHKVVKATN---FIGRVGHSELLRLLHRADAA
. : *
MSMEG_5641|M.smegmatis_MC2_155 VFPIQWEEPFGMVMIEAMACGTPVVALRGGAVPEVIVDGVTGVICDVP--
TH_0092|M.thermoresistible__bu LFPIQWEEPFGMVMIEAMACGTPVVALRGGAVPEVIVDGVTGVICEDP--
MAV_1024|M.avium_104 LFPIQWEEPFGIVMIEAMVCGTPVVALRGGAVSEVVADGVTGVVCDHP--
MLBr_00452|M.leprae_Br4923 CAPNIGGESFGIVLVEAMAAGTPVVASDLDAFRRVLRDGEVGHLVPAG--
MUL_3252|M.ulcerans_Agy99 CAPNTGGESFGIVLVEAMAAGTPVVASDLDAFRRVLRDGEIGRLVPVD--
MAB_2894c|M.abscessus_ATCC_199 CAPNIGGESFGIVLVEAMAAGTAVVASSLDAFRRVLLDGTAGRLTPTG--
Mb1244c|M.bovis_AF2122/97 VCPSVY-EPLGIVNLEAMACATAVVASDVGGIPEVVADGITGSLVHYD--
Rv1212c|M.tuberculosis_H37Rv VCPSVY-EPLGIVNLEAMACATAVVASDVGGIPEVVADGITGSLVHYD--
MMAR_4226|M.marinum_M VCASIY-EPLGIVNLEAMACATAVVASDVGGIPEVVADGITGTLVHYA--
Mflv_0483|M.gilvum_PYR-GCK LVTPLWDEPYGQVVVEAMACGTPVVAFAGGGIPELV-DEHSGRLVEPGDV
Mvan_2096|M.vanbaalenii_PYR-1 VLPSHY-EPFGIVALEAAATGTPLVTSTAGGLGEAVIDGVTGLSYPPG--
. *. * * :** . .*.:*: ... . : * *
MSMEG_5641|M.smegmatis_MC2_155 EELPAAMEKATHLD-PAACRRHVAAHFGVGRFGAGYEQIYRNVLR----Q
TH_0092|M.thermoresistible__bu AELPAAIERTADMD-PAECRRHVAANFDIERFGSGYEQIFRGVVDGSMAV
MAV_1024|M.avium_104 DELPGAIARAQTLD-PHACRRHVAANFAVAQFGSAYEAIYRNVLQHATAA
MLBr_00452|M.leprae_Br4923 DSAALADALVALLR-NDVLRERYVAAGAEAVRRYDWSVVASQIMRVYETV
MUL_3252|M.ulcerans_Agy99 DSDALAAALIEVLD-SQELRERYVAAGTEAVRRYDWSVVASQIMRVYETV
MAB_2894c|M.abscessus_ATCC_199 DPDALAAALIDVLG-NDEGRAQLVAAGTEAVQRYDWSVVAQQIMLVYETV
Mb1244c|M.bovis_AF2122/97 ADDATG--YQARLA-EAVNALVADPATAERYGHAGRQRCIQEFSWAYIAE
Rv1212c|M.tuberculosis_H37Rv ADDATG--YQARLA-EAVNALVADPATAERYGHAGRQRCIQEFSWAYIAE
MMAR_4226|M.marinum_M ADDPAG--YQSRLA-QAVNALVADPAKAERYGQAGRQRCIEEFSWTQIAE
Mflv_0483|M.gilvum_PYR-GCK AALAAAVREAVALP-RAGVRRVAVQRHGLARMVTGYVALYRALIAEATPP
Mvan_2096|M.vanbaalenii_PYR-1 DVAALARAVRAVLDSPAAAQRRARAARERLTSDFSWHTVAAETAQVYLAA
. . :
MSMEG_5641|M.smegmatis_MC2_155 RTRMTLREVLT--EPPGVA----ERASA---------
TH_0092|M.thermoresistible__bu RSNVALRQILP--ETGERD----EQIA----------
MAV_1024|M.avium_104 PRRMRSTGLVESIETGRRACDFRQKAPA---------
MLBr_00452|M.leprae_Br4923 ATSGSKVQVAS--------------------------
MUL_3252|M.ulcerans_Agy99 AGAGVKVQVAS--------------------------
MAB_2894c|M.abscessus_ATCC_199 TASGARVKVDSW-------------------------
Mb1244c|M.bovis_AF2122/97 QTLDIYRKVCA--------------------------
Rv1212c|M.tuberculosis_H37Rv QTLDIYRKVCA--------------------------
MMAR_4226|M.marinum_M QTLDIYRKVCR--------------------------
Mflv_0483|M.gilvum_PYR-GCK PRPPKLHSSSESANQSQAVSATARACLTNVSGVSAET
Mvan_2096|M.vanbaalenii_PYR-1 KRAVRRPQPRRPIEERPLPDR----------------