For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VFIQRHAVVRGIFGTFAPAEGTTLMKILIVSWEYPPVVIGGLGRHVHHLSTALAAAGHDVVVLSRRPSGT DPCTHPTSDEISEGVRVIAAAQDPHEFTFSNDMMAWTLAMGHAMIRTGLSLTRHSSDLPWRPDVVHAHDW LVAHPAITLAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVHAVESWLVRESDSLITCSASMCNEIIELF GPGLAEITVIRNGIDPARWPFAARRARTGPAELLYVGRLEYEKGVHDVIAALPRIRRSYPGTTLTIAGEG TQQDWLVDQARKYKVIKATRFVGHLNHNELLAALQRADAAVLPSHYEPFGLVALEAAAAGTPLVTSNIGG LGEAVINGQTGVSCPPRDIAELAAMVCTVLEDPDAAQQRALAARERLTSDFDWQTVAQQTAQVYLAAKRR ERQPQPRLPIVEHALPDR
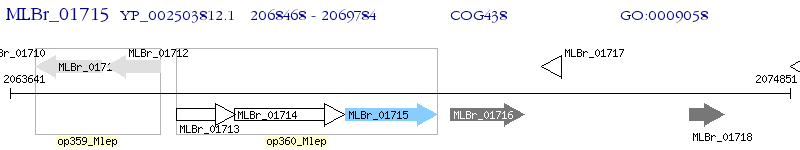
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01715 | - | - | 100% (438) | putative transferase |
| M. leprae Br4923 | MLBr_02443 | - | 2e-23 | 27.09% (395) | putative glycosyl transferase |
| M. leprae Br4923 | MLBr_00452 | - | 9e-19 | 25.33% (383) | putative glycosyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3058 | - | 0.0 | 87.92% (414) | transferase |
| M. gilvum PYR-GCK | Mflv_4262 | - | 0.0 | 74.64% (414) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv3032 | - | 0.0 | 87.92% (414) | transferase |
| M. abscessus ATCC 19977 | MAB_3366 | - | 1e-178 | 72.77% (415) | glycosyltransferase |
| M. marinum M | MMAR_1681 | - | 0.0 | 84.06% (414) | transferase |
| M. avium 104 | MAV_3879 | - | 0.0 | 84.45% (418) | glycosyl transferase, group 1 family protein |
| M. smegmatis MC2 155 | MSMEG_5080 | - | 2e-28 | 28.44% (422) | glycogen synthase |
| M. thermoresistible (build 8) | TH_2191 | - | 0.0 | 77.16% (416) | POSSIBLE TRANSFERASE |
| M. ulcerans Agy99 | MUL_1919 | - | 0.0 | 83.82% (414) | transferase |
| M. vanbaalenii PYR-1 | Mvan_2096 | - | 0.0 | 76.03% (413) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3058|M.bovis_AF2122/97 ------------------------MRILMVSWEYPPVVIGGLGRHVHHLS
Rv3032|M.tuberculosis_H37Rv ------------------------MRILMVSWEYPPVVIGGLGRHVHHLS
MLBr_01715|M.leprae_Br4923 MFIQRHAVVRGIFGTFAPAEGTTLMKILIVSWEYPPVVIGGLGRHVHHLS
MAV_3879|M.avium_104 ------------------------MKILMVSWEYPPVVIGGLGRHVHHLS
MMAR_1681|M.marinum_M ------------------------MKILMVSWEYPPVVIGGLGRHVHHLA
MUL_1919|M.ulcerans_Agy99 ------------------------MKILMVSWEYPPVVIGGLGRHVHHLA
Mflv_4262|M.gilvum_PYR-GCK ----------------------MKMKILLVSWEYPPVVIGGLGRHVHHLA
Mvan_2096|M.vanbaalenii_PYR-1 ----------------------MKTKILLVSWEYPPVVIGGLGRHVHHLA
TH_2191|M.thermoresistible__bu ------------------------MKILMVSWEYPPVVVGGLGRHVYHLS
MAB_3366|M.abscessus_ATCC_1997 ------------------------MRILMVSWEYPPVVIGGLGRHVHHLS
MSMEG_5080|M.smegmatis_MC2_155 ------------------------MRVAMMTREYPPEVYGGAGVHVTELV
:: ::: **** * ** * ** .*
Mb3058|M.bovis_AF2122/97 TALAAAGHDVVVLSRCPSGTDPSTHPSSDEVTEGVRVIAAAQDPHEFTFG
Rv3032|M.tuberculosis_H37Rv TALAAAGHDVVVLSRCPSGTDPSTHPSSDEVTEGVRVIAAAQDPHEFTFG
MLBr_01715|M.leprae_Br4923 TALAAAGHDVVVLSRRPSGTDPCTHPTSDEISEGVRVIAAAQDPHEFTFS
MAV_3879|M.avium_104 TALAAAGHDVVVLSRRPTGTDPRTHPSTDQISEGVRVIAAAQDPHEFTFG
MMAR_1681|M.marinum_M TALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVIAAAQDPHEFSFG
MUL_1919|M.ulcerans_Agy99 TALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVIAAAQDPHEFSFG
Mflv_4262|M.gilvum_PYR-GCK TELVAAGHEVVVLSRRPTGTDPQTHPTTDETHEGVRVIAAAEDPHEFGFG
Mvan_2096|M.vanbaalenii_PYR-1 TELAQSGHEVVVLSRRPTATDPQTHPTTDEVHEGVRVIAAAEDPHEFTFG
TH_2191|M.thermoresistible__bu TALAAAGHEVVVLSRRPTGTDPSTHPSTDEVRDGVRVLAAAQDPHEFDFG
MAB_3366|M.abscessus_ATCC_1997 TELAAAGHDVVVLTRRPSGTDPSSHPTSDEISEGVRVVAAAEDPHDFDFG
MSMEG_5080|M.smegmatis_MC2_155 AQLRKLCDVDVHCMGAPRDGAYVAHPDPTLRGANAALTMLSADLNMVNN-
: * . * * :** . .. : : * : .
Mb3058|M.bovis_AF2122/97 NDMMAWTLAMGHAMIRAGLR----LKKLGTDRSWRPDVVHAHDWLVAHPA
Rv3032|M.tuberculosis_H37Rv NDMMAWTLAMGHAMIRAGLR----LKKLGTDRSWRPDVVHAHDWLVAHPA
MLBr_01715|M.leprae_Br4923 NDMMAWTLAMGHAMIRTGLS----LTRHSSDLPWRPDVVHAHDWLVAHPA
MAV_3879|M.avium_104 TDMMAWTLAMGHAMIRAGLPPKLHDKKPGGNRPWRPDVVHAHDWLVAHPA
MMAR_1681|M.marinum_M SDMMAWTLAMGHAMVRAGLT----LSGPGAEQPWQPDIVHAHDWLVAHPA
MUL_1919|M.ulcerans_Agy99 SDMMAWTLAMGHAMVRAGLT----LSGPGAEQPWQPDIVHAHDWLVAHPA
Mflv_4262|M.gilvum_PYR-GCK TDMMAWVLAMGHSMIRAGFG----------LDGWRPDIVHAHDWLVAHPA
Mvan_2096|M.vanbaalenii_PYR-1 TDMMAWTLAMGHAMIRAGLV----------LADWRPDIVHAHDWLVATPA
TH_2191|M.thermoresistible__bu RDMMAWTLAMGHSMVRAGLALG--LRGGTGGTPWQPDVVHAHDWLVAHPA
MAB_3366|M.abscessus_ATCC_1997 TDMMAWTLAMGHAMVRAGLALG-----LKGSGDWRPDVVHAHDWLVAHPA
MSMEG_5080|M.smegmatis_MC2_155 --------------------------------AEAATVVHSHTWYTGLAG
. :**:* * .. ..
Mb3058|M.bovis_AF2122/97 IALAQFYDVPMVSTIHATEAGR-HSGWVSGALSRQVHAVESWLVRESDSL
Rv3032|M.tuberculosis_H37Rv IALAQFYDVPMVSTIHATEAGR-HSGWVSGALSRQVHAVESWLVRESDSL
MLBr_01715|M.leprae_Br4923 ITLAQFYDVPMVSTIHATEAGR-HSGWVSGALSRQVHAVESWLVRESDSL
MAV_3879|M.avium_104 IALAQFYDVPMVSTIHATEAGR-HSGWVSGPISRQVHAIESWLVRESDSL
MMAR_1681|M.marinum_M IALADYYDVPMVSTIHATEAGR-HSGWVSGDLSRQVHAVESWLVRESDSL
MUL_1919|M.ulcerans_Agy99 IALADYYDVPMVSTIHATEAGR-HSGWVSGDLSRQVHAVESWLVRESDSL
Mflv_4262|M.gilvum_PYR-GCK VALAQFFDVPLVSTIHATEAGR-HSGWVSGSVSRQVHALESWLVRDSDSL
Mvan_2096|M.vanbaalenii_PYR-1 VALAQFFDVPLVSTIHATEAGR-HSGWVSGQVSRQVHALESWLVRDSDSL
TH_2191|M.thermoresistible__bu IALAEFFDVPLVSTIHATEAGR-HSGWVSGPISRQVHAVESWLVRDSDSL
MAB_3366|M.abscessus_ATCC_1997 IALAEHFDVPLVSTLHATEAGR-HSGWVSGRISRQVHSVEWWLARESDSL
MSMEG_5080|M.smegmatis_MC2_155 HLASLLYGVPHVLTAHSLEPLRPWKAEQLGGGYQVSSWVERTAVEAADAV
: :.** * * *: *. * .. * : :* .. :*::
Mb3058|M.bovis_AF2122/97 ITCSASMNDEITELFGPG-LAEITVIRNGIDAARWPFAARRPRTG-----
Rv3032|M.tuberculosis_H37Rv ITCSASMNDEITELFGPG-LAEITVIRNGIDAARWPFAARRPRTG-----
MLBr_01715|M.leprae_Br4923 ITCSASMCNEIIELFGPG-LAEITVIRNGIDPARWPFAARRARTG-----
MAV_3879|M.avium_104 ITCSASMADEITELFGPG-LAQITVIRNGIETARWPFAPRRPRTG-----
MMAR_1681|M.marinum_M ITCSGSMSDEITELFGPG-LAETAVIRNGIDAARWPFAVRKPRTG-----
MUL_1919|M.ulcerans_Agy99 ITCSGSMSDEITELFGPG-LAETAVIRNGIDVARWPFAVRKPRTG-----
Mflv_4262|M.gilvum_PYR-GCK ITCSASMRDEISALFGPE-LAEISVIPNGIDTDGWPFAARRAHSG-----
Mvan_2096|M.vanbaalenii_PYR-1 ITCSASMRDEITTLFGPE-LADITVIPNGIDTDGWPFAPHRPRPG-----
TH_2191|M.thermoresistible__bu ITCSASMSEEITELFGPG-LSEIRVIRNGIDVAQWPFARRSPRSG-----
MAB_3366|M.abscessus_ATCC_1997 ITCSASMLDEVTQLFGPE-LPQVHVIRNGIDVTRWVFAPRPPADGT----
MSMEG_5080|M.smegmatis_MC2_155 IAVSSGMRDDVLRTYPALDPDRVHVVRNGIDTTVWYPAEPGPDESVLAEL
*: *..* ::: : . *: ***: * * . .
Mb3058|M.bovis_AF2122/97 -----PAELLYVGRLEYEKGVHDAIAALPRLRRTHPGTTLTIAGEGTQQD
Rv3032|M.tuberculosis_H37Rv -----PAELLYVGRLEYEKGVHDAIAALPRLRRTHPGTTLTIAGEGTQQD
MLBr_01715|M.leprae_Br4923 -----PAELLYVGRLEYEKGVHDVIAALPRIRRSYPGTTLTIAGEGTQQD
MAV_3879|M.avium_104 -----PAELLYLGRLEYEKGVHDVIAALPRIRRTHPGTTLTIAGEGTQQG
MMAR_1681|M.marinum_M -----PPELLYVGRLEYEKGVHNAIAALPRIRRAHPGTTLTIAGEGTQQD
MUL_1919|M.ulcerans_Agy99 -----PPELLYVGRLEYEKGVHNAIAALPRIRRAHPGTTLTIAGEGTQQD
Mflv_4262|M.gilvum_PYR-GCK -----PAELLYFGRLEYEKGVHDAIAALPKVRRTHPGTTLTIAGDGTQLG
Mvan_2096|M.vanbaalenii_PYR-1 -----PAELLYFGRLEYEKGVHDLIAALPRIRRAHPGTTLTIAGDGTQLD
TH_2191|M.thermoresistible__bu -----PPRLLYVGRLEYEKGVQDAIAALPRIRRTHPGTTLTIAGTGTQHE
MAB_3366|M.abscessus_ATCC_1997 -----PPVLLFVGRLEYEKGIHDAIAALPKIRRAHPGTVLAVAGDGTQQD
MSMEG_5080|M.smegmatis_MC2_155 GVDLNRPIVAFVGRITRQKGVAHLVAAAHRFAPDVQLVLCAGAPDTPQIA
. : :.**: :**: . :** :. . : * .*
Mb3058|M.bovis_AF2122/97 WLIDQARKHRVLRAT---RFVGHLDHTELLALLHRADAAVLPSHYEPFGL
Rv3032|M.tuberculosis_H37Rv WLIDQARKHRVLRAT---RFVGHLDHTELLALLHRADAAVLPSHYEPFGL
MLBr_01715|M.leprae_Br4923 WLVDQARKYKVIKAT---RFVGHLNHNELLAALQRADAAVLPSHYEPFGL
MAV_3879|M.avium_104 WLIEQARKHRVLKAI---RFVGHLEHSELLAMLHRADAAVLPSHYEPFGI
MMAR_1681|M.marinum_M WLVEQARKHRVLKAT---KFLGHLEHAELLTLLHRADAAVFPSHYEPFGL
MUL_1919|M.ulcerans_Agy99 WLVEQARKHRVLKAT---KFLGHLEHAELLTLLHRADAAVFPSHYEPFGL
Mflv_4262|M.gilvum_PYR-GCK FLTEQARRHKVLKAT---KFVGRVDHSQLLTLLHRADVAVLPSHYEPFGI
Mvan_2096|M.vanbaalenii_PYR-1 FLVEQARKHKVVKAT---NFIGRVGHSELLRLLHRADAAVLPSHYEPFGI
TH_2191|M.thermoresistible__bu WLVEQARKHKVLKAT---RFVGQLGHEALLRQLHTADAAVLPSHYEPFGI
MAB_3366|M.abscessus_ATCC_1997 WLLEQARKYKVVKSV---QFLGTLDHSELLHWLHHADAILLPSHYEPFGI
MSMEG_5080|M.smegmatis_MC2_155 EEVSSAVQQLAQARTGVFWVREMLPTHKIREILSAATVFVCPSVYEPLGI
..* : . . : : * * . : ** ***:*:
Mb3058|M.bovis_AF2122/97 VALEAAAAGTPLVTSNIGGLGEAVINGQTGVSCAPRD------VAGLAAA
Rv3032|M.tuberculosis_H37Rv VALEAAAAGTPLVTSNIGGLGEAVINGQTGVSCAPRD------VAGLAAA
MLBr_01715|M.leprae_Br4923 VALEAAAAGTPLVTSNIGGLGEAVINGQTGVSCPPRD------IAELAAM
MAV_3879|M.avium_104 VALEAAAAGTPLVTSNIGGLGEAVIDGQTGVSCPPRD------VPALAAA
MMAR_1681|M.marinum_M VALEAAAAGTPLVTSNIGGLGEAVIDGVTGVSCPPRD------VAALAAA
MUL_1919|M.ulcerans_Agy99 VALEAAAAGTPLVTSNIGGLGEAVIDGVTGVSCPPRD------VAALAAA
Mflv_4262|M.gilvum_PYR-GCK VALEAAATGTPLVTSTAGGLGEAVIDGVTGMSYPPSD------VIALAAA
Mvan_2096|M.vanbaalenii_PYR-1 VALEAAATGTPLVTSTAGGLGEAVIDGVTGLSYPPGD------VAALARA
TH_2191|M.thermoresistible__bu VALEAAATGAPLVTSTAGGLGEAVIDGVTGKSFKPRD------IAGLTAA
MAB_3366|M.abscessus_ATCC_1997 VALEAAAAGTPLVTSTVGGLGEAVIDGETGMSFAPRD------VAGIAEA
MSMEG_5080|M.smegmatis_MC2_155 VNLEAMACATAVVASDVGGIPEVVADGRTGLLVHYDANDTEAYEARLAEA
* *** * .:.:*:* **: *.* :* ** ::
Mb3058|M.bovis_AF2122/97 VRSVLDDPAAAQRRARAARQRLTSDFDWQTVATATAQVYLAAKRGERQPQ
Rv3032|M.tuberculosis_H37Rv VRSVLDDPAAAQRRARAARQRLTSDFDWQTVATATAQVYLAAKRGERQPQ
MLBr_01715|M.leprae_Br4923 VCTVLEDPDAAQQRALAARERLTSDFDWQTVAQQTAQVYLAAKRRERQPQ
MAV_3879|M.avium_104 IRTVLDDPEAAQRRACAARERLTSDFDWHTVADKTAQVYLSAKRAERQPQ
MMAR_1681|M.marinum_M VCGVLDDPAAAQQRARAARERLNSDFDWQTVAEQTAQVYLAAKRGERQPL
MUL_1919|M.ulcerans_Agy99 VCGVLDDPAAAQQRARAARERLNSNFDWQTVAEQTAQVYLAAKRGERQPL
Mflv_4262|M.gilvum_PYR-GCK IRAVLDAPAAAQERATAARARLTSDFAWHTVAAETAQVYLAAKRAERRPQ
Mvan_2096|M.vanbaalenii_PYR-1 VRAVLDSPAAAQRRARAARERLTSDFSWHTVAAETAQVYLAAKRAVRRPQ
TH_2191|M.thermoresistible__bu VRAVLDDPEGAQQRAVAARERLTADFDWHSVAAETAQVYLAAKRREREPL
MAB_3366|M.abscessus_ATCC_1997 VTRTLDDPAAAAERAVAARARLTADFDWATVADETAQVYLAAKRREREPL
MSMEG_5080|M.smegmatis_MC2_155 VNSLVADPDRAREYGVAGRERCIEEFSWAHIAEQTLEIYRKVSA------
: : * * . . *.* * :* * :* * ::* ..
Mb3058|M.bovis_AF2122/97 PRLPIVEHALPDR
Rv3032|M.tuberculosis_H37Rv PRLPIVEHALPDR
MLBr_01715|M.leprae_Br4923 PRLPIVEHALPDR
MAV_3879|M.avium_104 ARLPIVEHALPDR
MMAR_1681|M.marinum_M PRLPIVEHALPDR
MUL_1919|M.ulcerans_Agy99 PRLPIVEHALPDR
Mflv_4262|M.gilvum_PYR-GCK PRRTIVERPLPDR
Mvan_2096|M.vanbaalenii_PYR-1 PRRPIEERPLPDR
TH_2191|M.thermoresistible__bu PRRLIVEHALPDR
MAB_3366|M.abscessus_ATCC_1997 GRSVIVERPLPDR
MSMEG_5080|M.smegmatis_MC2_155 -------------