For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KYRGPLIGLSLFMIVAITLTWLVYVTLRRDVAGEKVPYSAMFTDVFGLREGDDVRMAGVRVGRVEKIELD GKLAKVSFVVQGDQQVLGTTVASVTYQNIVGQRYLGLTLGSQGEPVPLPAGSVLPLERSDPSFDVTTLLN GYEPLFSLLDPQAADDLTQGVIQSLQGDEASLVALVDQTSQLTESFAGHDEELGAVITDLNTVMSNLAKH NDSMDQVLTETQAAVSIFDAQRQELVDSTGSISRVVSRLSTVSDDVYPSLNELIEREPGFAQHLVSIEPQ LAFTGANLPLLLKGFARITSEGAYANAYACDLNATGFFPGLNDVTPIIVDAATPGNDAQYSPKCRNMANG *
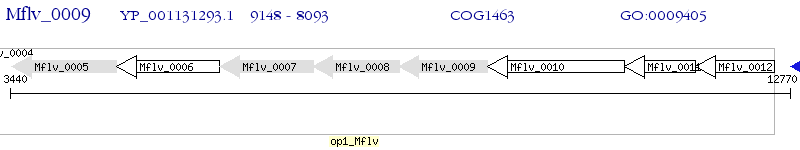
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0009 | - | - | 100% (351) | virulence factor Mce family protein |
| M. gilvum PYR-GCK | Mflv_0710 | - | 2e-41 | 31.60% (326) | virulence factor Mce family protein |
| M. gilvum PYR-GCK | Mflv_4161 | - | 3e-37 | 30.99% (313) | virulence factor Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0605 | mce2B | 8e-39 | 31.90% (326) | MCE-family protein MCE2B |
| M. tuberculosis H37Rv | Rv0590 | mce2B | 1e-35 | 33.99% (253) | MCE-family protein MCE2B |
| M. leprae Br4923 | MLBr_02590 | mce1B | 9e-43 | 30.58% (327) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_1009c | - | 1e-140 | 69.03% (352) | putative MCE family protein |
| M. marinum M | MMAR_3865 | mce5B | 1e-139 | 66.95% (351) | Mce family protein, Mce5B |
| M. avium 104 | MAV_2387 | - | 1e-135 | 65.24% (351) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_1144 | - | 1e-158 | 76.07% (351) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_3156 | - | 1e-155 | 74.36% (351) | PUTATIVE virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_3797 | mce5B | 1e-138 | 66.67% (351) | Mce family protein, Mce5B |
| M. vanbaalenii PYR-1 | Mvan_0960 | - | 1e-171 | 84.62% (351) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0009|M.gilvum_PYR-GCK MKYRGPLIGLSLFMIVAITLTWLVYVTLRRDVAGEKVPYSAMFTDVFGLR
Mvan_0960|M.vanbaalenii_PYR-1 MKYRGALVGLSLFMVVAVTLTWLVYVTLRRDVAGSTAPYAAVFTDVFGLR
MSMEG_1144|M.smegmatis_MC2_155 MKIRGALIGLSLFMVVALTLTWMVYVTLRRDVAGRTVPYTAVFTDVFGLR
TH_3156|M.thermoresistible__bu MRSRGALIGLSLFMAVAVVLTWLVYVTLRRDVAGSTVPYSAVFTDVFGLR
MMAR_3865|M.marinum_M MKFRGPLIGLSLFMVVAVALTWMVYVSLRREVAGKTVAYAAVFTDVYGLR
MUL_3797|M.ulcerans_Agy99 MKFRGPLIGLSLFMVVAVALTWMVYVSLRREVAGKTVAYAAVFTDVYGLR
MAV_2387|M.avium_104 MRFRGPLIALTLFMIVSLTLTWLVYVSLRRDVAGDTARYSAVFSDVYGLR
MAB_1009c|M.abscessus_ATCC_199 MKFRGPLIALTVFMAVALALTWLVYATLRRDVAGGTTSYSAIFTDVYGLR
Mb0605|M.bovis_AF2122/97 MKTTGTTIKLGIVWLVLSVFTVMIIVVFGQVRFHHTTGYSAVFTHVSGLR
Rv0590|M.tuberculosis_H37Rv MKTTGTTIKLGIVWLVLSVFTVMIIVVFGQVRFHHTTGYSAVFTHVSGLR
MLBr_02590|M.leprae_Br4923 MKLTGTVVRLSIFSLVLLLFTVMIIVVFGQMRFGRTNGYTAEFTNISGLR
*: *. : * :. * :* :: . : : . *:* *:.: ***
Mflv_0009|M.gilvum_PYR-GCK EGDDVRMAGVRVGRVEKIELDG--KLAKVSFVVQGDQQVLGTTVASVTYQ
Mvan_0960|M.vanbaalenii_PYR-1 EGDDVRMAGVRVGRVEKIELQG--DLAKVSFVVQNDQQVLGTTVASVTYQ
MSMEG_1144|M.smegmatis_MC2_155 EGDDVRMAGVRVGRVEKIELDG--KHAKVSFVVQADQQVFGNTVASVTYQ
TH_3156|M.thermoresistible__bu EGDDVRMAGVRVGRVERIELDG--KYAKVSFVVQSDQQVLGNTVASVTYQ
MMAR_3865|M.marinum_M EGDDVRMAGVRVGRVEKIDLEG--KLAKVRFVVQNEQRLFGNTVASVTYQ
MUL_3797|M.ulcerans_Agy99 EGDGVRMAGVRVGRVEKIDLEG--KLAKVRFVVQNEQRLFGNTVASVTYQ
MAV_2387|M.avium_104 EGDDVRMAGVRVGRVEKIELDG--KLAKVSFVVQSEQRLYGNTLASVTYQ
MAB_1009c|M.abscessus_ATCC_199 DGDDVRMAGVRVGRVEKIELVNN-KQAKVDFVVQNDQKLYGNTLASVTYQ
Mb0605|M.bovis_AF2122/97 AGQFVRAAGVEVGKVAKVTLIDGDKQVLVDFTVDRSLSLDQATTASIRYL
Rv0590|M.tuberculosis_H37Rv AGQFVRAAGVEVGKVAKVTLIDGDKQVLVDFTVDRSLSLDQATTASIRYL
MLBr_02590|M.leprae_Br4923 TGQFVRASGVEVGKVNSVALINGGTRVRVKFNVDRSVPLYQSTTAQIRYL
*: ** :**.**:* : * . . * * *: . : * *.: *
Mflv_0009|M.gilvum_PYR-GCK NIVGQRYLGLTLGS-QGEPVPLPAGSVLPLERSDPSFDVTTLLNGYEPLF
Mvan_0960|M.vanbaalenii_PYR-1 NIVGQRYLGLSLGN-TGDPRPLPAGSVIPLERSEASFDVTTLLNGYEPLF
MSMEG_1144|M.smegmatis_MC2_155 NIVGQRYLGLSLGK-EGDPTLLAAGSVIPLERTDPSFDVGTLLNGYEPLF
TH_3156|M.thermoresistible__bu NIVGQRYLGLSLGN-LGDPHPLPPGATIPVEQTDPSFDVGALLNGYEPLF
MMAR_3865|M.marinum_M NIVGQRYLGLSLGK-KGSPDQLAPGSVIPVQRTEPSFDVTALLNGYEPLF
MUL_3797|M.ulcerans_Agy99 NIVGQRYLGLSLGK-KGSPDQLAPGSVIPVQRTEPSFDVTALLNGYEPLF
MAV_2387|M.avium_104 NIVGQRYLGLSLGK-EGNPAQLPPGSTIPLERTEPSFDVTTLLNGYEPLF
MAB_1009c|M.abscessus_ATCC_199 NIVGQRYLGLSLGK-TGDTNVLAGGSTIPVERTDPSFDVTTLLNGYEPLF
Mb0605|M.bovis_AF2122/97 NLIGDRYLELGRG---HSGQRLAPGATIPLEHTHPALDLDALLGGFRPLF
Rv0590|M.tuberculosis_H37Rv NLIGDRYLELGRG---HSGQRLAPGATIPLEHTHPALDLDALLGGFRPLF
MLBr_02590|M.leprae_Br4923 DLIGNRYLELKHGEGQGSEKIMPAGGLIPLSRTSPALDLDALIGGFKPLF
:::*:*** * * . :. *. :*:.:: .::*: :*:.*:.***
Mflv_0009|M.gilvum_PYR-GCK SLLDPQAADDLTQGVIQSLQGDEASLVALVDQTSQLTESFAGHDEELGAV
Mvan_0960|M.vanbaalenii_PYR-1 SLLNPRDADNLTQGVIQSLQGDEASIVGLVDQTSQLTESLAGRDEELGTV
MSMEG_1144|M.smegmatis_MC2_155 SVLDPKYANNLTEGVIQSLQGDQGSITALIDQTAQLTEAFAGREEELGSV
TH_3156|M.thermoresistible__bu SVLDPKHADNLTKGVIESLQGDEASITALVDQTAQLTEAFAGRDEELGAV
MMAR_3865|M.marinum_M SLLNPRDADNLTKGIIESLQGDTASLATLVSQTSTLTETFAGRDQALGDV
MUL_3797|M.ulcerans_Agy99 SLLNPRDADNLTKGIIESLQGDTASLATLVSQTSTLTETFAGRDQALGDV
MAV_2387|M.avium_104 SLLNPHDADNLTKGIIASLQGDTSSLATLVGQTSTLTQTFAGRDQALGNV
MAB_1009c|M.abscessus_ATCC_199 STLSPEQADNLTKGVIQSLQGDQASITSLVDQTSTLTKTFAGRDQVLGNV
Mb0605|M.bovis_AF2122/97 QTLDPDKVNSIASSIITVFQGQGATINDILDQTASLTATLADRDHAIGEV
Rv0590|M.tuberculosis_H37Rv QTLDPDKVNSIASSIITVFQGQGATINDILDQTASLTATLADRDHAIGEV
MLBr_02590|M.leprae_Br4923 RALDPDKINTIASAIVAAFQGQGGTINDILDQTAQLTTAIGERDQAIGEV
*.* : ::..:: :**: .:: ::.**: ** ::. ::. :* *
Mflv_0009|M.gilvum_PYR-GCK ITDLNTVMSNLAKHNDSMDQVLTETQAAVSIFDAQRQELVDSTGSISRVV
Mvan_0960|M.vanbaalenii_PYR-1 ITDLNAVMGNLARHNDSLDQVLTQVQSAVTTFDGQRQELVDSTGSMARVV
MSMEG_1144|M.smegmatis_MC2_155 ITDLNAVAANLARHNDDLDHIVSEAQSVVATFEARRPELVESMGSIAKVV
TH_3156|M.thermoresistible__bu ITDLNTVVANLARHNDDLDHVIGEARSVVSTFDDRRDQLIDSMGSAAKVI
MMAR_3865|M.marinum_M ITNLNKVIYNLAQQNDNLDGVITDTREVVSTLDQRRPELVSSIGSLARVL
MUL_3797|M.ulcerans_Agy99 ITNLNKVIYNLAQQNDNLDGVITDTREVVSTLDQRRPELVSSIGSLARVL
MAV_2387|M.avium_104 ITNLNKVVGNLAAQNDNLDGVITQTRSVVGELDRRRPDLVASVGSLARLS
MAB_1009c|M.abscessus_ATCC_199 ITSLSTVTGNLAQQNDSLDKAITNTRKVVADFDSRRPELVNSVGSLAQTM
Mb0605|M.bovis_AF2122/97 VNNLNTVLATTVKHQTEFDRTVDKLEVLITGLKNRADPLAAAAAHISSAA
Rv0590|M.tuberculosis_H37Rv VNNLNTVLATTVKHQTEFDRTVDKLEVLITGLKNRADPLAAAAAHISSAA
MLBr_02590|M.leprae_Br4923 VKNLNVVLDTTVRHRKEFDQTVNNLEILITGLKDHGDQLAGSFAHISNAA
:..*. * . . :. .:* : . . : :. : * : . :
Mflv_0009|M.gilvum_PYR-GCK SRLSTVSDDVYPSLNELIEREPGFAQHLVSIEPQLAFTGANLPLLLKGFA
Mvan_0960|M.vanbaalenii_PYR-1 RQLSTISDEVYPSLNELITREPGFTQHMVSIEPQLAFTGANLPLLLKGFA
MSMEG_1144|M.smegmatis_MC2_155 RQLSTISDEVYPSLSEMVQRQPGFAAHMLGIEPQLAFVGANLPLMLKGFA
TH_3156|M.thermoresistible__bu RQLSAISDEVYQPLNEMVQREPGFAKHMLGIEPQLAFVGANLPLMLKGFA
MMAR_3865|M.marinum_M ERLSASANDVYPQLREFIDRQPGATRHILSVEPQVAFFGDNIPLLLKGLT
MUL_3797|M.ulcerans_Agy99 ERLSASANDVYPQLREFIDRQPGATRHILSVEPQVAFFGDNIPLLLKGLT
MAV_2387|M.avium_104 DRLSASAADVYPALREFIDRQPGVTKHIMDVEPQVAFFGDNIPLLLKGLT
MAB_1009c|M.abscessus_ATCC_199 RRLSKITDEVYPSLDELITRQPGFAKHMVQIEPKLAFLGGNLPLLLKGLA
Mb0605|M.bovis_AF2122/97 GTLADLLGADRPLLHSSFGHLEGIQQPLIDELAELDHVLGKLPDAYRIIG
Rv0590|M.tuberculosis_H37Rv GTLADLLG--------------------------------------RIVH
MLBr_02590|M.leprae_Br4923 GTVANLLAEDHALLHKSINYLDGIQQPLIDQRLQLEDYLHKLPTVLNQIG
:: . .
Mflv_0009|M.gilvum_PYR-GCK RITSE-GAYANAYACDLNATGFFPGLNDVTPIIVDAATPGNDAQYSPKCR
Mvan_0960|M.vanbaalenii_PYR-1 RITGE-GAYANAYACDLNIQGFFPGLNDVVPIIVDAATPGNDARYTPKCR
MSMEG_1144|M.smegmatis_MC2_155 RITQE-GTYANAYLCDLNATAFFPGLNDVTPIIVDAATPGNKAQHTPKCR
TH_3156|M.thermoresistible__bu RVTGE-GTYANAYACNLDGTAFFPGLNDITPIIVAAATPGNKARYTPKCR
MMAR_3865|M.marinum_M RVGNR-GAYGNAYVCDINAFGFFPGLNDVMPIIVNAATPGNQARHTPRCR
MUL_3797|M.ulcerans_Agy99 RVGNR-GAYGNAYVCDINAFGFFPGLNDVMPIIVNAATPGNQARHTPRCR
MAV_2387|M.avium_104 RVGNQ-GAYGNAYVCDVNFMGFFPGLNDVVPIIINAATPGNRAWHTPRCR
MAB_1009c|M.abscessus_ATCC_199 RITGD-GAYGNAYICDLNITGFFPGLNDVVPIIVNAATPGNVAQHTPRCR
Mb0605|M.bovis_AF2122/97 RAGGIYGDFFNFYLCDI--SLKVNGLQPGGPVRTVKLFGQPTGRCTPQ--
Rv0590|M.tuberculosis_H37Rv CCT------------------AASGTSRASSSRS----------------
MLBr_02590|M.leprae_Br4923 RTIGSYGDFVNFYSCDI--TLKINGLQPGGPVRTVRLFQQPTGRCTPQ--
* . .
Mflv_0009|M.gilvum_PYR-GCK NMANG
Mvan_0960|M.vanbaalenii_PYR-1 NMANG
MSMEG_1144|M.smegmatis_MC2_155 SMANG
TH_3156|M.thermoresistible__bu NLANG
MMAR_3865|M.marinum_M SMADG
MUL_3797|M.ulcerans_Agy99 SMADG
MAV_2387|M.avium_104 STVDG
MAB_1009c|M.abscessus_ATCC_199 NLANG
Mb0605|M.bovis_AF2122/97 -----
Rv0590|M.tuberculosis_H37Rv -----
MLBr_02590|M.leprae_Br4923 -----