For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAGAGRSTFELTKALVNAQLRSDHEYRNLVGSVQAPHCILNTRISRNRRFATQQYPLDRLKAIGAQYDAT INDVALAIIGGGLRRFLDELGELPNKSLIVVLPVNVRPKDDEGGGNAVATILATLGTDVADPVQRLAAVT ASTRAAKAQLRSMDKDAILAYSAALMAPYGVQLASTLSGVKPPWPYTFNLCVSNVPGPEDVLYLRGSRME ASYPVSLVAHSQALNVTLQSYAGTLNFGFIGCRDTLPHLQRLAVYTGEALDQLAAADGAAGLGS
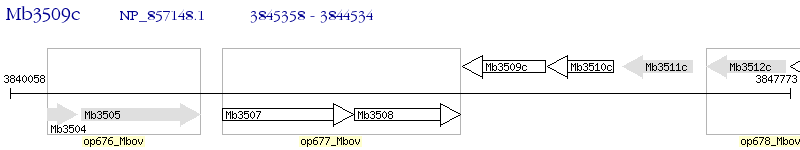
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3509c | - | - | 100% (274) | hypothetical protein Mb3509c |
| M. bovis AF2122 / 97 | Mb3766c | - | 3e-48 | 39.78% (269) | hypothetical protein Mb3766c |
| M. bovis AF2122 / 97 | Mb3761c | - | 9e-48 | 39.63% (270) | hypothetical protein Mb3761c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1058 | - | 1e-102 | 66.92% (266) | hypothetical protein Mflv_1058 |
| M. tuberculosis H37Rv | Rv3480c | - | 1e-154 | 100.00% (274) | hypothetical protein Rv3480c |
| M. leprae Br4923 | MLBr_01244 | - | 8e-07 | 28.23% (209) | hypothetical protein MLBr_01244 |
| M. abscessus ATCC 19977 | MAB_4544c | - | 6e-44 | 38.30% (235) | hypothetical protein MAB_4544c |
| M. marinum M | MMAR_4955 | - | 1e-133 | 88.59% (263) | hypothetical protein MMAR_4955 |
| M. avium 104 | MAV_0355 | - | 2e-45 | 38.89% (270) | bifunctional wax ester synthase/acyl-coadiacylglycerol |
| M. smegmatis MC2 155 | MSMEG_6322 | - | 2e-47 | 43.28% (238) | bifunctional wax ester synthase/acyl-CoA diacylglycerol |
| M. thermoresistible (build 8) | TH_2030 | - | 2e-44 | 38.29% (269) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2706 | - | 2e-45 | 40.23% (261) | hypothetical protein MUL_2706 |
| M. vanbaalenii PYR-1 | Mvan_5773 | - | 1e-101 | 67.69% (260) | hypothetical protein Mvan_5773 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3509c|M.bovis_AF2122/97 --------------------------------------------------
Rv3480c|M.tuberculosis_H37Rv MSQTARRLGPQDMF----FLYSESSTTMMHVGALMPFTPP-SGAPPDLLR
MMAR_4955|M.marinum_M MSKTAKRLGPQDMF----FLYSESPSTMMHVGALMPFTPP-DDAPADFLR
Mflv_1058|M.gilvum_PYR-GCK MSANSVRLGLQDLM----FIYGETSSSKMHVGGLLPFTPP-ADAPRDYLR
Mvan_5773|M.vanbaalenii_PYR-1 -------------M----FIYGETSSSKMHVAGLLPFTPP-ADAPRDYLR
MUL_2706|M.ulcerans_Agy99 ----MKLLSPLDQM----FARMEAPRTPMHIGAFAIFDMP-KNAPRGFIR
MSMEG_6322|M.smegmatis_MC2_155 -MNRMQLMSPTDSM----FLIAESREHPMHVGGLALYDPP-DDAGPEFVR
TH_2030|M.thermoresistible__bu ----MRLISPADSM----FLLAESREHPMHVAGLQLFEPPPGTAGPEFLQ
MAV_0355|M.avium_104 ----MELIMPTDAV----FLLGESREHPMHVGGLQLYQPP-EGAGRNFVR
MAB_4544c|M.abscessus_ATCC_199 ----MPFMPVSDSM----FLIGETREHPFHVGGLQLFKPP-VDAGPDYAE
MLBr_01244|M.leprae_Br4923 MADSVEGIGPFDELGALDYLLHRGEANPRTRAGIMAVELLDTTPDWNRFR
Mb3509c|M.bovis_AF2122/97 --------------------------------------------------
Rv3480c|M.tuberculosis_H37Rv QLVDESKASEVVEPWSLRLSHPELLYH-PTQSWVVDDNFDLDYHVRRSAL
MMAR_4955|M.marinum_M QLVDESKANEVVEPWNLKLSHPDLLYS-PTQSWVYDENFDLDYHVRRSAL
Mflv_1058|M.gilvum_PYR-GCK GMIDEARRQEVVAPWNRRLAYPRLQFS-PLHSWVTDEDFDFDYHVRRSAL
Mvan_5773|M.vanbaalenii_PYR-1 RMVDDARRQEVMPPWNRKLAYPRLQFS-PLHTWVTDTEFDFDYHVRRSAL
MUL_2706|M.ulcerans_Agy99 DLYEAVSQLAFLP-----FPFDSVIVGGPSMAYWKQVQPDPSSHVRMSAL
MSMEG_6322|M.smegmatis_MC2_155 ELYEEMVRHTDFQPVFRKHPAT-LLGGIANVGWTLDDEVDLDYHLRRSAL
TH_2030|M.thermoresistible__bu GLHQRLAAGRELHPTFRKHPAT-LFGGIAHLGWAVDDDVDFDYHLRRSAL
MAV_0355|M.avium_104 DFYKQLVAQPDFQPTFRKHPAK-FVGGIANLGWSYDADVDVDYHVRRSAL
MAB_4544c|M.abscessus_ATCC_199 VFYEQLMSTTEVSSDFRKRPGQ-PLAVMGNLTWIVDDEVDWEYHVRRSAL
MLBr_01244|M.leprae_Br4923 SRIEDVSQR----VLRLRQKVVVPTLPTAAPRWVVDPDFELDFHVRRVRV
Mb3509c|M.bovis_AF2122/97 --------------------------------------------------
Rv3480c|M.tuberculosis_H37Rv ASPGDERELGIPVSRLHSHALDLRRPPWEVHFIEGLEGGRFAIYIKMHHS
MMAR_4955|M.marinum_M ASPGDERELGILVSRLHSHALDLRRPPWEMHFIEGLEGGRFAIYIKMHHS
Mflv_1058|M.gilvum_PYR-GCK ASPGDERELGVLVSRLHSNHLDLTRPPWELHVIEGLEGGRFALYMKIHHA
Mvan_5773|M.vanbaalenii_PYR-1 ASPGDERELGILVSRLHSNHLDLTRPPWELHVIEGLEGGRFALYMKIHHA
MUL_2706|M.ulcerans_Agy99 PQPGTARDLGALVERLHSTPLDMTKPLWELHVIEGLEGKQFAIYFKAHHC
MSMEG_6322|M.smegmatis_MC2_155 PSPGRPRELLELTSRVHGTLLDRHRPLWEAYLIEGMADGRFAVYTKVHHS
TH_2030|M.thermoresistible__bu PGPGRIRELLDLTSRWHGTLLDRHRPLWEVHLVEGLADGRVAIYFKVHHA
MAV_0355|M.avium_104 PSPRRVRELLELTSRWHSSLLDRHRPLWETHIVEGLKDGRFAIYTKVHHA
MAB_4544c|M.abscessus_ATCC_199 PRPARVRELLTVTSRWHSSPLDRHRPLWEMHVVEGLSDGRLAVYTKMHHA
MLBr_01244|M.leprae_Br4923 PDPGTLREVFDLAEVIQQSPMDVSRPLWTATLVEGLAAGRAAMLLQISHA
Mb3509c|M.bovis_AF2122/97 --------------------------------------------------
Rv3480c|M.tuberculosis_H37Rv LIDGYTGQKMLARSLSTDPHDTTHPLFFNIPTPGRSPADTQDSVGGGLIA
MMAR_4955|M.marinum_M LIDGYTGQKMLARSLSADPHDTTHPLFFNIPTPGPAPADTEESAGGGLIG
Mflv_1058|M.gilvum_PYR-GCK LVDGYSAMRMLGRSLSTDPASRDTRMFFNVPSPTRSRRD----------P
Mvan_5773|M.vanbaalenii_PYR-1 LVDGYTAMRMLGRSLSTDPETRDARMFFNVPIPKRSRPT----------G
MUL_2706|M.ulcerans_Agy99 AVDGMGGVNLIKSWLTTDPTAP--------PGSGKPEP-----------L
MSMEG_6322|M.smegmatis_MC2_155 LIDGVSAMKLVERTLSEDPSDTTVRVPWNLPRRESSRR-----------A
TH_2030|M.thermoresistible__bu LIDGVAVMKLMQRTLSTDPGDD-ARVPWNLPPPRRNP------------A
MAV_0355|M.avium_104 LIDGVSAQKLMQRALSSDPDDPEIRAPWTLP--KRSRK-----------A
MAB_4544c|M.abscessus_ATCC_199 VIDGVGALRMMMRSLSDDPDARDCQAVWAPAKRRAKSS-----------I
MLBr_01244|M.leprae_Br4923 ITDGVGSVEMFAEMYDLERDPPSRPRSPQPIPQDLSRND---------LM
Mb3509c|M.bovis_AF2122/97 -----------------------------MAGAGRSTFELTKALVNAQLR
Rv3480c|M.tuberculosis_H37Rv GAGNVLDGLGDVVRGLGGLVSGVGSVLGSVAGAGRSTFELTKALVNAQLR
MMAR_4955|M.marinum_M GAGNVLSGLGDVVKGLGSMVSGVGSVLGSVASAGRSSLDLTRALVNSQLR
Mflv_1058|M.gilvum_PYR-GCK GAAESSNPLTATLRALGGVSS-------AVTGGVSSAVDLTNALVNTQIR
Mvan_5773|M.vanbaalenii_PYR-1 AGAQSSNPVTATLRALGAVGS-------TVSDGVGSAVDLASALVNTQIR
MUL_2706|M.ulcerans_Agy99 GDDYDLATVFAATTAKR------------AVEGVSAVSELIGKIISMALG
MSMEG_6322|M.smegmatis_MC2_155 GSSS------LARTATG------------AATSLAALAPSTIRLARAALL
TH_2030|M.thermoresistible__bu GPVS------RLRSATG------------AVGSVAALAPSTLSLARAALL
MAV_0355|M.avium_104 GPSS------RLSSLVH------------AAGSVAALAPSTVSLARAALV
MAB_4544c|M.abscessus_ATCC_199 VSTTNTSAIDLVKGAAG------------AVRQVVSIPPGLLKYGRHALS
MLBr_01244|M.leprae_Br4923 LQGINHLPVALAGGVVGG---------LSGVASVAGRAILRPASTVSGVV
. :
Mb3509c|M.bovis_AF2122/97 SDHEYRNLVGSVQAPHCILNTRISRNRRFATQQYPLDRLKAIGAQYDATI
Rv3480c|M.tuberculosis_H37Rv SDHEYRNLVGSVQAPHCILNTRISRNRRFATQQYPLDRLKAIGAQYDATI
MMAR_4955|M.marinum_M TDNEYRDLVSSVQAPHCILNTRISRNRRFATQQYPLARMKAIGAQYDATV
Mflv_1058|M.gilvum_PYR-GCK RDGENAHIAGSVSAPHSILNARISRNRRFATQQYEFDRLKKLSSQHGATL
Mvan_5773|M.vanbaalenii_PYR-1 RDGDFGRISGSASAPHSILNARISRNRRFATQQYEFDRLKKLSSQHGATI
MUL_2706|M.ulcerans_Agy99 AN---SSVRAALTTPGTPFNTRINRHRRLAVQVLSLPRLKAVSSATGTTI
MSMEG_6322|M.smegmatis_MC2_155 EQ----QLTLPFGAPRTMFNVKIGGARRVAAQSWPLERLRRIKAVTGATI
TH_2030|M.thermoresistible__bu EQ----RLTLPFGAPRTMFNVRIGGARRVAAQSWPLQRIRRIRQAAGXTL
MAV_0355|M.avium_104 EQ----QLTLPFGAPRTMLNVKIGGARRCAAQSWPLERIKNVKNAAGVTV
MAB_4544c|M.abscessus_ATCC_199 DP----QLVKPLSAPHTMLNVSIGGARRFAAQTWSMARIKEAGKALGGTV
MLBr_01244|M.leprae_Br4923 GYVRSGIRVLSQAAEPSPLLRQRSLATRTEAIEIQLSDLHKAAKAGDGSI
. : : . * . : :: . ::
Mb3509c|M.bovis_AF2122/97 NDVALAIIGGGLRRFLDELGELPNKSLIVVLPVNVRPKDDEGGG-NAVAT
Rv3480c|M.tuberculosis_H37Rv NDVALAIIGGGLRRFLDELGELPNKSLIVVLPVNVRPKDDEGGG-NAVAT
MMAR_4955|M.marinum_M NDVAMAIIGGGLRRFLDELGELPDKSLIAVLPVNVRPKDDEGGG-NAVAT
Mflv_1058|M.gilvum_PYR-GCK NDVALAIIGGGLRKFLSDFDKLPDRSLIAFLPVNVRPKGDEGGG-NAVGA
Mvan_5773|M.vanbaalenii_PYR-1 NDVALAIIGGGLRKFLADLGELPDRSLIAFLPVNVRPKGDEGGG-NAVGA
MUL_2706|M.ulcerans_Agy99 NDVVLASIGGACRHYLRDQSALPKSNLTVSVPVGFER--DADTV-NAASG
MSMEG_6322|M.smegmatis_MC2_155 NDIVLAMCAGALRAYLAEQDALPDRPLIAMVPVSMRSEHEADAGGNMVGS
TH_2030|M.thermoresistible__bu NDVALAMCAGALRQYLLEHDALPATPLVAMVPVSLRSADESAGGGNRTGL
MAV_0355|M.avium_104 NDVVLAMCSGALRYYLLEQNALPDTPLIAMVPVSLRTEEEADAGGNLVGA
MAB_4544c|M.abscessus_ATCC_199 NDVVLAMCGGALRTYLEEQNALPDKPLVAMCPVSIRAEGDQNAG-NSITA
MLBr_01244|M.leprae_Br4923 NDAYLAGLVGALRRYHEALG-VSISTLPMAVPVNVRTEADVVGS-NRFVG
** :* *. * : . :. * **... : *
Mb3509c|M.bovis_AF2122/97 ILATLGTDVADPVQRLAAVTASTRAAKAQLRSMDKDAILAYSAALMAPYG
Rv3480c|M.tuberculosis_H37Rv ILATLGTDVADPVQRLAAVTASTRAAKAQLRSMDKDAILAYSAALMAPYG
MMAR_4955|M.marinum_M ILATLGTDIADPVERLRAVTASTRMAKAQLKNMDRDAILAYSAALMVPYG
Mflv_1058|M.gilvum_PYR-GCK ILAPMGTDIEDPVERLDTITTATRAAKGQLQSMSPAAIIAYSAALLAPAG
Mvan_5773|M.vanbaalenii_PYR-1 ILAPMGTDIEDPVERLAVITAATRASKAQLQSMSPAAIIAYSAALLAPAG
MUL_2706|M.ulcerans_Agy99 FVTQLGTSVEDPIERLTTISASTSRGKAELLALSPNALQHYSVFGLLPIA
MSMEG_6322|M.smegmatis_MC2_155 ILCSLGTDVEDPADRLAVIRRSITDNKRVFSELPRLQALALSALLIAPLG
TH_2030|M.thermoresistible__bu VLCNLATDSDDPAERLDRISSSMRSNKRVFSQLPRVQAMALSAAMIAPLG
MAV_0355|M.avium_104 ILCNLATDSDDPAQRLLTISESMRSNKKVFSQLPRFQALALSAANLSSLA
MAB_4544c|M.abscessus_ATCC_199 LLANLATDKPDPLERFSAIRDSVQAAKSVLSEMTPLQRMLVGALNGAPVL
MLBr_01244|M.leprae_Br4923 VTLAAPLGTNDPAARMQKIRSQMTQRR--------DEPAMNIIGSLAPLM
. . ** *: : : .
Mb3509c|M.bovis_AF2122/97 VQLASTLSGVKPPWPYTFNLCVSNVPGPEDVLYLRGSRMEASYPVS----
Rv3480c|M.tuberculosis_H37Rv VQLASTLSGVKPPWPYTFNLCVSNVPGPEDVLYLRGSRMEASYPVS----
MMAR_4955|M.marinum_M IQLASTLTGVKPPLPYTFNLCVSNVPGPQEVLYLRGSRMEASYPVS----
Mflv_1058|M.gilvum_PYR-GCK SQIAGALTGVQPPWPYTFNLCVSNVPGPREPLYFNGSRLEATYPVS----
Mvan_5773|M.vanbaalenii_PYR-1 GQIAGALTGVNPPWPHTFNLCVSNVPGPREPLYFNGSRLEATFPVS----
MUL_2706|M.ulcerans_Agy99 VGQKSGALGIIPPL---FNFTVSNVVLSKEPLYLSGAKLDVIVPMS----
MSMEG_6322|M.smegmatis_MC2_155 LTT--AFPGFVDATAPPFNLVISNVPGPKKPLYWRGARLAGNYPLS----
TH_2030|M.thermoresistible__bu LG---AVPGVVSSAPPPFNLVISNVPGAPEPRYWMGARLAGNYPLS----
MAV_0355|M.avium_104 LA---AVPGWVSATSPPFNIVISNVPGPTAPLYYGGARLDGNYPLS----
MAB_4544c|M.abscessus_ATCC_199 TG---AVPGLVDRTAPAFNVIISNVPGPRTDMFWNGFEMDGCYPAS----
MLBr_01244|M.leprae_Br4923 TVLPASVLDFIVDSVASSDVNASNIPAYPGDTYFAGAKILRQYGIGPRPG
. :. **: : * .: .
Mb3509c|M.bovis_AF2122/97 ------LVAHSQALNVTLQSYAGTLNFGFIGCRDTLPHLQRLAVYTGEAL
Rv3480c|M.tuberculosis_H37Rv ------LVAHSQALNVTLQSYAGTLNFGFIGCRDTLPHLQRLAVYTGEAL
MMAR_4955|M.marinum_M ------LVAHSQALNVTLQSYAGTLNFGFIGCRDTLPHLQRLAVYTGEAL
Mflv_1058|M.gilvum_PYR-GCK ------IPIHGMALNITLQSYADTMNLGFVGCRDRLPHLQRLAVYTGDAL
Mvan_5773|M.vanbaalenii_PYR-1 ------IPIHGMALNITLQSYADTMNLGFVGCRDRLPRLQRLAVYTGDAL
MUL_2706|M.ulcerans_Agy99 ------FLCDGYGLNVTLVGYTDKVVLGFLGCRDTLPHLQRLARYTGEAF
MSMEG_6322|M.smegmatis_MC2_155 ------IALDGQALNMTVVSNAHNLDFGLVGCRRSVPHLQRLLGHLETSL
TH_2030|M.thermoresistible__bu ------IVLDGQALNITLVTNADNLDFGLVGARGSVPHLQRLLGHLEDSL
MAV_0355|M.avium_104 ------IVLDGQALNITLASNAGNLDFGLVGCRRSVPHLQRLLMHLESSL
MAB_4544c|M.abscessus_ATCC_199 ------IPIDGVALNITCTSSGSSMHFGLTGCRTSVPHLQRILAHLEDAL
MLBr_01244|M.leprae_Br4923 VAMMAVLMSRGGFCTVTVRYDRASVKSEALFARCLLEGFDEVLALAGDPT
: . .:* .: .* : ::.: .
Mb3509c|M.bovis_AF2122/97 DQLAAADGAAGLGS---------
Rv3480c|M.tuberculosis_H37Rv DQLAAADGAAGLGS---------
MMAR_4955|M.marinum_M DELDEPAAASAVN----------
Mflv_1058|M.gilvum_PYR-GCK AELEAASQI--------------
Mvan_5773|M.vanbaalenii_PYR-1 GELEAASS---------------
MUL_2706|M.ulcerans_Agy99 DELETATTTA-------------
MSMEG_6322|M.smegmatis_MC2_155 KDLETATGA--------------
TH_2030|M.thermoresistible__bu QQLERAVGG--------------
MAV_0355|M.avium_104 KDLERAVGE--------------
MAB_4544c|M.abscessus_ATCC_199 TQLEESVA---------------
MLBr_01244|M.leprae_Br4923 PHAVPASFAARSSGSPAGWLSSS
. .