For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LACVRRSCDVTGTARAGIGAGADPAVVDAVAVAADDCGFATLWVGEHVVMVDRPASRYPYSRDGVIAVPA QADWLDPMIALSFAAAASSRVDVATGVLLLPEHNPVIVAKEAASLDRLSGRRLTLGVASDGPRRSSTRSE CHSSGAQSAPPNTSLQCAHYGATTSHRSTATVGS
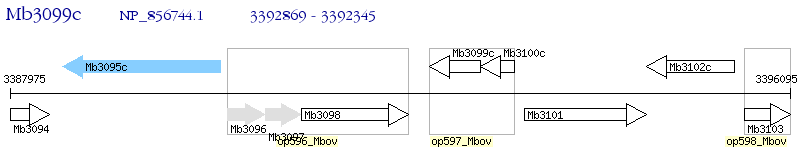
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3099c | - | - | 100% (174) | hypothetical protein Mb3099c |
| M. bovis AF2122 / 97 | Mb0978c | - | 3e-12 | 36.45% (107) | oxidoreductase |
| M. bovis AF2122 / 97 | Mb2185c | - | 2e-11 | 39.22% (102) | hypothetical protein Mb2185c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4839 | - | 3e-33 | 62.75% (102) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3072c | - | 3e-97 | 100.00% (174) | hypothetical protein Rv3072c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0366 | - | 3e-19 | 43.52% (108) | hypothetical protein MAB_0366 |
| M. marinum M | MMAR_1586 | - | 6e-50 | 81.90% (116) | oxidoreductase |
| M. avium 104 | MAV_2935 | - | 1e-17 | 41.28% (109) | hypothetical protein MAV_2935 |
| M. smegmatis MC2 155 | MSMEG_2811 | - | 1e-43 | 76.58% (111) | hydride transferase 1 |
| M. thermoresistible (build 8) | TH_2830 | - | 2e-14 | 41.24% (97) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_3448 | - | 6e-49 | 83.64% (110) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_4864 | - | 5e-14 | 40.19% (107) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0366|M.abscessus_ATCC_1997 -------MKIGVVAPVELGVTAEPDKILEFARAAERLGFSEISVVEHAVV
MAV_2935|M.avium_104 ------------MAPVADGVTADPDWMVAFARHLEACGFESIVAVEHTVL
Mb3099c|M.bovis_AF2122/97 MACVRRSCDVTGTARAGIGAGADPAVVDAVAVAADDCGFATLWVGEHVVM
Rv3072c|M.tuberculosis_H37Rv MACVRRSCDVTGTARAGIGAGADPAVVDAVAVAADDCGFATLWVGEHVVM
MMAR_1586|M.marinum_M --------MQLGLHALGIGAGADRSMIDAVACAADDAGFATLWAGEHVVM
MUL_3448|M.ulcerans_Agy99 --------MQLGLHALEIGAGADRSMIDAVACAADDAGFATLWAGEHVVM
MSMEG_2811|M.smegmatis_MC2_155 --------MQISLHALGIGAGARREIIDAVAVAAERAGFTTLWCGEHVVM
Mflv_4839|M.gilvum_PYR-GCK -------------------------MIDAVAASAERSGFETLWFGDQVVM
TH_2830|M.thermoresistible__bu -----------MDFGLVLFTSDRGITPAKAATLADDHGFTTFYVPEHTHI
Mvan_4864|M.vanbaalenii_PYR-1 -----------MNYGLVLFTSDRGITPAAAAKLADEHGFETFYVPEHTHI
* : ** : ::. :
MAB_0366|M.abscessus_ATCC_1997 IGDTQSTYPYSPTGQSHLPDDIDIPDPLELLSFVAAATTTLGLSTGVLVL
MAV_2935|M.avium_104 LTRYDSVYPYDSSGRVGLAPDCPIPDPLDLLAFLAGHTTRLGLATGVLVL
Mb3099c|M.bovis_AF2122/97 VDRPASRYPYSRDGVIAVPAQADWLDPMIALSFAAAASSRVDVATGVLLL
Rv3072c|M.tuberculosis_H37Rv VDRPASRYPYSRDGVIAVPAQADWLDPMIALSFAAAASSRVDVATGVLLL
MMAR_1586|M.marinum_M VDRHTSRYPYSEDGVIAIPPQADWLDPMIALSFAAAASSRITVATGVLLL
MUL_3448|M.ulcerans_Agy99 VDRHTSRYPYSEDGVIAIPPQADWLDPMIALSFAAASSSRITVATGVLLL
MSMEG_2811|M.smegmatis_MC2_155 VDRPRSRYPYSADGRIAVAADADWLDPMIALSFAAAATSTIRIATGVLLL
Mflv_4839|M.gilvum_PYR-GCK ADEDGPHHPYRDDGEIPASAQTDWLDPLIALSFAAAATNRIELATGVLVL
TH_2830|M.thermoresistible__bu PVKREAAHPTTGDSSLPDDRYMRTLDPWVSLATASAVTSRVRLSTAVALP
Mvan_4864|M.vanbaalenii_PYR-1 PIKRQAAHPTTGDESLPDDRYMRTLDPWVSLGTAAAVTSRVRLSTAVALP
. :* ** *. :. :. : ::*.* :
MAB_0366|M.abscessus_ATCC_1997 PDHHPVVLAKRLATLDRLSRGRLRICAGVGWMREEIEACGGDFDRRGRAA
MAV_2935|M.avium_104 PNHHPVVLAKRVGTLDALSGGRLRLCVGVGWLQEELRACGADFDNRGRRA
Mb3099c|M.bovis_AF2122/97 PEHNPVIVAKEAASLDRLSGRRLTLGVAS---------------------
Rv3072c|M.tuberculosis_H37Rv PEHNPVIVAKEAASLDRLSGRRLTLGVAS---------------------
MMAR_1586|M.marinum_M PEHNPVVVAKQAASLDRLSGGRLTLGVGVGWSREEFAALGVPFQDRAGRT
MUL_3448|M.ulcerans_Agy99 PEHNPVVVAKQAASLDRLSGGRLTLGVGVGWSREEFAALGVPFQDRVGRT
MSMEG_2811|M.smegmatis_MC2_155 PEHNPVIMAKQAASVDQLSGGRLLLGVGIGWSREEFEALGVPFARRGART
Mflv_4839|M.gilvum_PYR-GCK PERNPVVTAKQAASLDRMTGGRLTLGVGIGISPQEYDALGVPFASRGARL
TH_2830|M.thermoresistible__bu VEHDPISLAKTIASLDHLSGGRVSLGVGFGWNTDELVDHGVPPKRRRTML
Mvan_4864|M.vanbaalenii_PYR-1 VEHDPITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHNVPPGRRRTML
::.*: ** .::* :: *: : ..
MAB_0366|M.abscessus_ATCC_1997 DEAIAVMRALWST--DGPTSHDGEFYSFRNAYSYPKPVRPQGVPLYIGGH
MAV_2935|M.avium_104 DEQLAVLRALWAHRPDG-ASFHGEFFTFDNVMCYPKPVAAERFPVHIGGH
Mb3099c|M.bovis_AF2122/97 --------------------------------------------------
Rv3072c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1586|M.marinum_M AEYVAAMRTLWRD---DLASFTGEFVRFDSVRVNPKPVRDRRVPVMVGGN
MUL_3448|M.ulcerans_Agy99 AEYVAAMRTLWRD---DLASFTGEFVRFDSVRVNPKPVRDRRVPVMVGGN
MSMEG_2811|M.smegmatis_MC2_155 AEYVEAMRTLWRQ---DVADFTGEFAAFDAVRLHPKPSAG-SIPIIVGGN
Mflv_4839|M.gilvum_PYR-GCK AEYVGAMRTLWRD---DDASFAGRYVAFDGVRANPKPQGR-SIPVVLGGN
TH_2830|M.thermoresistible__bu REYLEAMRALWTQ---EEASYQGEFVSFGPSWAWPKPVQAHIPVLVGAAG
Mvan_4864|M.vanbaalenii_PYR-1 REYLEAMRALWTQ---EEAEFDGEFVKFGPSWAWPKPVQAHIPVLVGAAG
MAB_0366|M.abscessus_ATCC_1997 SKAAARRAGRLGSGFQPLGLVGDD-LTAAVGVMRAAAVDAGRDPRGLDLV
MAV_2935|M.avium_104 SRAAARRAGRFGDGFQPLGVTGAR-LASLIGLMRDEAVAAGRDPAALEVS
Mb3099c|M.bovis_AF2122/97 --------------------DGP-----RRSSTRSECHSSGAQS------
Rv3072c|M.tuberculosis_H37Rv --------------------DGP-----RRSSTRSECHSSGAQS------
MMAR_1586|M.marinum_M SDAALRRVAAWADGWYGFNLDGVGAVRERVGTLDRLCAESGRNRDELRLS
MUL_3448|M.ulcerans_Agy99 SDAALRRVAAWADGWYGFNLDGVGAVRERVGTLDRLCAESGRNRDELRLS
MSMEG_2811|M.smegmatis_MC2_155 GDRALARVASWGDGWYGFNLDGVAAVGERMDMLRRLCARAGRDVDDLHCA
Mflv_4839|M.gilvum_PYR-GCK SDAALRRVAEWGDGWYGFDLADVEEAAFRASKLRSMCREFGRDPTELKLS
TH_2830|M.thermoresistible__bu TEKNFKWIARSADGWITTPRDVD--IEEPVKLLQDIWADAGRDGAPQIVA
Mvan_4864|M.vanbaalenii_PYR-1 TEKNFKWIAKSADGWITTPRDFT--IDEPVKLLQDTWAAAGRDGAPQIVA
* :
MAB_0366|M.abscessus_ATCC_1997 LGHGLHRVDQAALDQAAHAGAGRMLLSASRRATTLEAVLEEMAGCATRLE
MAV_2935|M.avium_104 LGHSVSKIDAERAARLADQGADRLVLAMPP-TSDIEQAKDMLSACAQRLS
Mb3099c|M.bovis_AF2122/97 ----------------------------APPNTSLQCAHYGATTSHRSTA
Rv3072c|M.tuberculosis_H37Rv ----------------------------APPNTSLQCAHYGATTSHRSTA
MMAR_1586|M.marinum_M V--ALRCPQPCDVDALAELGIDELVLVETPPEAADAAQNWVSALAERWLS
MUL_3448|M.ulcerans_Agy99 V--ALRCPQPCDVDALAELGIDELVLVETPPEAADAAQNWVSALAERWLS
MSMEG_2811|M.smegmatis_MC2_155 V--ALQAPAVGDIPALVDLGVDELVIVESPPQDPDVAADWVDALADRWIT
Mflv_4839|M.gilvum_PYR-GCK V--ALHDPVPADAVRLADAGIDELVLVAGPPADPIAAEAWIADLAGQWMR
TH_2830|M.thermoresistible__bu LD---FKPDPDKLARWRDLGVTEVLFGLPDRSDDEIAAYVERLAGKLGLS
Mvan_4864|M.vanbaalenii_PYR-1 LD---FKPDPEKLAHWREIGVTEVLFGLPDKSEADVAAYVERLAGKLGAL
MAB_0366|M.abscessus_ATCC_1997 LG---GPR
MAV_2935|M.avium_104 LRPVSGP-
Mb3099c|M.bovis_AF2122/97 TVGS----
Rv3072c|M.tuberculosis_H37Rv TVGS----
MMAR_1586|M.marinum_M TVWVR---
MUL_3448|M.ulcerans_Agy99 TVWVR---
MSMEG_2811|M.smegmatis_MC2_155 CI------
Mflv_4839|M.gilvum_PYR-GCK VRT-----
TH_2830|M.thermoresistible__bu --------
Mvan_4864|M.vanbaalenii_PYR-1 T-------