For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVITINRPEARNAVNGAVSIVVGDALEEAHDNPDVRAVVITGAGDKSLCAGADLKAIARRENPYHPHHGE WGIAGYRHHFIDKPTSAAVSGTALDDGAEPALASDLVVADEHT
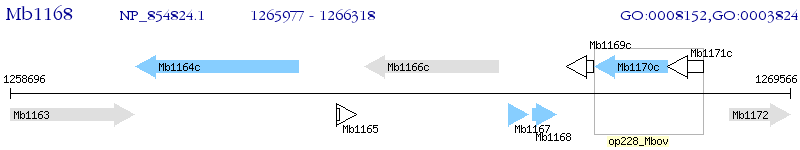
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1168 | - | - | 100% (113) | enoyl-CoA hydratase |
| M. bovis AF2122 / 97 | Mb2511 | echA14 | 7e-14 | 40.35% (114) | enoyl-CoA hydratase |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 8e-13 | 39.29% (112) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2117 | - | 7e-44 | 72.97% (111) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv1136 | - | 7e-63 | 100.00% (113) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 5e-11 | 37.50% (112) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_2058 | - | 8e-17 | 43.64% (110) | enoyl-CoA dehydratase/isomerase |
| M. marinum M | MMAR_4317 | echA1_1 | 3e-47 | 80.18% (111) | enoyl-CoA hydratase, EchA1_1 |
| M. avium 104 | MAV_1277 | - | 1e-46 | 81.98% (111) | carnitinyl-CoA dehydratase |
| M. smegmatis MC2 155 | MSMEG_5198 | - | 3e-41 | 72.07% (111) | carnitinyl-CoA dehydratase |
| M. thermoresistible (build 8) | TH_4237 | - | 1e-26 | 74.32% (74) | PUTATIVE carnitinyl-CoA dehydratase |
| M. ulcerans Agy99 | MUL_0972 | echA1_1 | 2e-45 | 78.38% (111) | enoyl-CoA hydratase, EchA1_1 |
| M. vanbaalenii PYR-1 | Mvan_4588 | - | 1e-42 | 72.07% (111) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1168|M.bovis_AF2122/97 --------------------MVITINRPEARNAVNGAVSIVVGDALEEAH
Rv1136|M.tuberculosis_H37Rv --------------------MVITINRPEARNAVNGAVSIVVGDALEEAH
MMAR_4317|M.marinum_M -MTDAGDDQPAGLVEHRGNVMVITINRPEARNAINGAVSVAVGDALESAQ
MUL_0972|M.ulcerans_Agy99 -MTDAGDDQPAGLVEHRGNVMVITINRPEARNAINGAVSVAVGGALESAQ
MAV_1277|M.avium_104 -----MTDAPGALAERRGNVMVITINRPEARNAVNAAVSIGVGDALEEAQ
Mflv_2117|M.gilvum_PYR-GCK -------MAPGALTERRGNVLIITINRPEARNAINASVSIAVGDALQAAQ
Mvan_4588|M.vanbaalenii_PYR-1 MTDTVTDTAPGALTERRGNVLIITINRPEARNAVNASVSIGVGNALQAAQ
MSMEG_5198|M.smegmatis_MC2_155 ----MTETAPGALTERRGNVLLITINRPEARNAINSSVSQAVGDALAEAQ
TH_4237|M.thermoresistible__bu --------------------------------------------------
MAB_2058|M.abscessus_ATCC_1997 ------MAEQPIRYEVVDSVAWLTINRPEARNALNNAVRTGLIDAVLRFN
MLBr_02402|M.leprae_Br4923 ------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAAKELD
Mb1168|M.bovis_AF2122/97 DNPDVRAVVITGAGDKSLCAGADLKAIARRENPYHPHHGEWGIAGYRHHF
Rv1136|M.tuberculosis_H37Rv DNPDVRAVVITGAGDKSLCAGADLKAIARRENPYHPHHGEWGIAGYRHHF
MMAR_4317|M.marinum_M NDPEVRAVVITGAGDKSFCAGADLLAISRRENIYHPDHNEWGFAGYVHHF
MUL_0972|M.ulcerans_Agy99 NDAEVRAVVITGAGDKSFCAGADLLAISRRENIYHPDHNEWGFAGYVHHF
MAV_1277|M.avium_104 HDPEVRAVVLTGAGDKSFCAGADLKAIARRENLYHPDHPEWGFAGYVRHF
Mflv_2117|M.gilvum_PYR-GCK EDPDVRAVILTGAGDKSFCAGADLKAISRGENLFHPEHSEWGFAGYVRHF
Mvan_4588|M.vanbaalenii_PYR-1 DDPEIRAVILTGAGDKSFCAGADLKAISRGENLFHPEHTEWGFAGYVRHF
MSMEG_5198|M.smegmatis_MC2_155 NDPEVRAVVLTGAGDKSFCAGADLKAISRGENLFHPEHPEYGFAGYVSHF
TH_4237|M.thermoresistible__bu -------VVLTGSGDKSFCAGADLKAIARGENLFHPDHPEWGFAGYVRHF
MAB_2058|M.abscessus_ATCC_1997 DDDAAKVLVLTGVGDKAFCAGGDLKEMAQNALKVPPKDFAPQFG--RNID
MLBr_02402|M.leprae_Br4923 IDPDVGAILITGS-PKVFAAGADIKEMAS-LTFTDAFDADFFSAWGKLAA
:::** * :.**.*: :: . . .
Mb1168|M.bovis_AF2122/97 IDKPTSAAVSGTALDDGAEPALASDLVVADEHT-----------------
Rv1136|M.tuberculosis_H37Rv IDKPTSAAVSGTALDDGAEPALASDLVVADEHT-----------------
MMAR_4317|M.marinum_M IDKPTIAAVNGTALGGGTELALASDLVVADERAKFGLPEVKRGLIAAAGG
MUL_0972|M.ulcerans_Agy99 IDKPTIAAVNGTALCGGTELALASDLVVADERAKFGLPEAKRGFIAAAGG
MAV_1277|M.avium_104 IDKPTIAAVNGTALGGGTELALASDLVVADERAQFGLPEVKRGLIAAAGG
Mflv_2117|M.gilvum_PYR-GCK IDKPTIAAVNGTALGGGTELALASDLVVAEERAKFGLPEVKRGLIAGAGG
Mvan_4588|M.vanbaalenii_PYR-1 IDKPTIAAVNGTALGGGTELALASDLVVAEERAKFGLPEVKRGLIAGAGG
MSMEG_5198|M.smegmatis_MC2_155 IDKPTIAAVNGTALGGGTELALASDLVVAEERAMFGLPEVKRGLIAGAGG
TH_4237|M.thermoresistible__bu MDKPTIAAVNGTALGGGTELALASDLVVARESAVFGLPEVKRGLIAGAGG
MAB_2058|M.abscessus_ATCC_1997 VAKPTIAAVNGVAFAGGFLLAQQCDLVVAAEHATFAVSEVKVG--RGSPW
MLBr_02402|M.leprae_Br4923 VRTPMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGG
: .* *** * *: .* * .**::* : :
Mb1168|M.bovis_AF2122/97 --------------------------------------------------
Rv1136|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4317|M.marinum_M VFRIVNQLPRKVAMEMLLTGEPITASDAYEWGLINQVVKQGAVLDAALAL
MUL_0972|M.ulcerans_Agy99 VFRIVNQLPRKVAMEMLLTGEPITASDAYEWGLINQVVKQGAVLDAALAL
MAV_1277|M.avium_104 VFRIAEQLPRKVAMRLLLTGEPLSAAAARDWGLINEVVEAGSVLDAALAL
Mflv_2117|M.gilvum_PYR-GCK VFRIVEQLPRKVALELIYTGEPISSADALRWGLINQVVPDGTVVDAALAL
Mvan_4588|M.vanbaalenii_PYR-1 VFRIVEQLPRKVALELIYTGEPISSADALRWGLINQVVPDGTVVDAALAL
MSMEG_5198|M.smegmatis_MC2_155 VFRIIEQLPRKVALELVYTGDPISAADALKWGLINQVVPDGTVVDAALAL
TH_4237|M.thermoresistible__bu VFRIVEQLPRKVALELLFTGEPITAAEALRWGLINEVVPDGAELDAALAL
MAB_2058|M.abscessus_ATCC_1997 ATPLSWLVPPRVAMQILLTGDPITAARAHQVGLVNEVVPADQLRERTQQL
MLBr_02402|M.leprae_Br4923 SQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAV
Mb1168|M.bovis_AF2122/97 --------------------------------------------------
Rv1136|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4317|M.marinum_M AARVTVNAPLSVRASKRIAYGVDDGVVADEERGWERTMREMRSLIRSEDA
MUL_0972|M.ulcerans_Agy99 AARVTVNAPLSVRASKRIAYGVDDGVVADEERGWERTMREMRSLIRSEDA
MAV_1277|M.avium_104 ASAITVNAPLSVQASKRIAYGVDDGVVVGDEPGWDRTMREMRALLKSEDA
Mflv_2117|M.gilvum_PYR-GCK AERITCNAPLAVRASKRVSYGAIDGVVGSDEPFWKQTFSEFSTLLKTEDA
Mvan_4588|M.vanbaalenii_PYR-1 AERITCNAPLAVQASKRVSYGAVDGVVGSEEPFWKQTFGEFSTLLKTEDA
MSMEG_5198|M.smegmatis_MC2_155 AERITCNAPLAVQASKRVAYGAEEGTVPAEQPSWKRTNREFVTLLKTEDA
TH_4237|M.thermoresistible__bu AERITVNAPLAVQASKRVAYGAENGTVPGDENGWKLTNREFSTLLKTEDA
MAB_2058|M.abscessus_ATCC_1997 ALSIAANAPLSVLASKRTVYLSAQHHLAA---AYDLADEIWEPVYLSDDA
MLBr_02402|M.leprae_Br4923 ATTISQMSRSATRMAKEAVNRSFESTLAEG---LLHERRLFHSTFVTDDQ
Mb1168|M.bovis_AF2122/97 ------------------
Rv1136|M.tuberculosis_H37Rv ------------------
MMAR_4317|M.marinum_M KEGPLAFAQKREPVWKAR
MUL_0972|M.ulcerans_Agy99 KEGPLAFAEKREPVWKTR
MAV_1277|M.avium_104 KEGPRAFAEKREPVWQAR
Mflv_2117|M.gilvum_PYR-GCK MEGPLAFAQKREPVWKAK
Mvan_4588|M.vanbaalenii_PYR-1 MEGPLAFAQKREPVWKAK
MSMEG_5198|M.smegmatis_MC2_155 MEGPLAFAQKRQPVWKAK
TH_4237|M.thermoresistible__bu KEGPLAFAEKREPVWKAR
MAB_2058|M.abscessus_ATCC_1997 QEGPAAFREKRAPQWKGH
MLBr_02402|M.leprae_Br4923 SEGMAAFIEKRAPQFTHR