For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAPGALTERRGNVLIITINRPEARNAINASVSIAVGDALQAAQEDPDVRAVILTGAGDKSFCAGADLKAI SRGENLFHPEHSEWGFAGYVRHFIDKPTIAAVNGTALGGGTELALASDLVVAEERAKFGLPEVKRGLIAG AGGVFRIVEQLPRKVALELIYTGEPISSADALRWGLINQVVPDGTVVDAALALAERITCNAPLAVRASKR VSYGAIDGVVGSDEPFWKQTFSEFSTLLKTEDAMEGPLAFAQKREPVWKAK
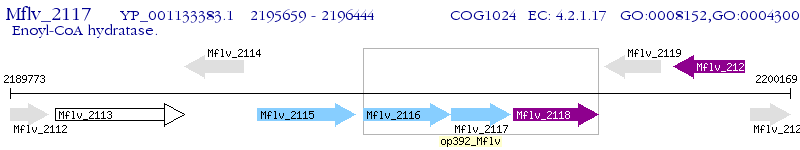
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2117 | - | - | 100% (261) | enoyl-CoA hydratase/isomerase |
| M. gilvum PYR-GCK | Mflv_4354 | - | 8e-57 | 46.90% (258) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_1542 | - | 2e-46 | 41.67% (264) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1168 | - | 2e-43 | 72.97% (111) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv0222 | echA1 | 5e-56 | 46.09% (256) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 3e-34 | 37.55% (245) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_4588c | - | 9e-57 | 47.51% (261) | enoyl-CoA hydratase/isomerase |
| M. marinum M | MMAR_4317 | echA1_1 | 1e-114 | 75.68% (259) | enoyl-CoA hydratase, EchA1_1 |
| M. avium 104 | MAV_2597 | - | 1e-119 | 78.99% (257) | carnitinyl-CoA dehydratase |
| M. smegmatis MC2 155 | MSMEG_5198 | - | 1e-129 | 87.69% (260) | carnitinyl-CoA dehydratase |
| M. thermoresistible (build 8) | TH_4237 | - | 1e-100 | 83.89% (211) | PUTATIVE carnitinyl-CoA dehydratase |
| M. ulcerans Agy99 | MUL_0972 | echA1_1 | 1e-110 | 72.97% (259) | enoyl-CoA hydratase, EchA1_1 |
| M. vanbaalenii PYR-1 | Mvan_4588 | - | 1e-142 | 95.77% (260) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0222|M.tuberculosis_H37Rv MSSESDAANTEPEVLVEQRDRILIITINRPKAKNAVNAAVSRGLADAMDQ
MAB_4588c|M.abscessus_ATCC_199 ---------MADEVLIEQRDRVLLITINRPDARNAVNRAVSQGLAAAADQ
MMAR_4317|M.marinum_M ---MTDAGDDQPAGLVEHRGNVMVITINRPEARNAINGAVSVAVGDALES
MUL_0972|M.ulcerans_Agy99 ---MTDAGDDQPAGLVEHRGNVMVITINRPEARNAINGAVSVAVGGALES
Mb1168|M.bovis_AF2122/97 ----------------------MVITINRPEARNAVNGAVSIVVGDALEE
Mflv_2117|M.gilvum_PYR-GCK --------MA-PGALTERRGNVLIITINRPEARNAINASVSIAVGDALQA
Mvan_4588|M.vanbaalenii_PYR-1 -MTDTVTDTA-PGALTERRGNVLIITINRPEARNAVNASVSIGVGNALQA
MSMEG_5198|M.smegmatis_MC2_155 -----MTETA-PGALTERRGNVLLITINRPEARNAINSSVSQAVGDALAE
TH_4237|M.thermoresistible__bu --------------------------------------------------
MAV_2597|M.avium_104 -MAETLTEVTRQAAVVERRGNVALITIDRPDARNAVNGAVSTAVGDALEE
MLBr_02402|M.leprae_Br4923 --------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAAKE
Rv0222|M.tuberculosis_H37Rv LDGDAGLSVAILTGGGG-SFCAGMDLKAFARGEN----VVVEGRGLGFTE
MAB_4588c|M.abscessus_ATCC_199 LDSSADLSVAIITGAGG-NFCAGMDLKAFVSGEA----VLSE-RGLGFTN
MMAR_4317|M.marinum_M AQNDPEVRAVVITGAGDKSFCAGADLLAISRRENIYHPDHNEWGFAGYVH
MUL_0972|M.ulcerans_Agy99 AQNDAEVRAVVITGAGDKSFCAGADLLAISRRENIYHPDHNEWGFAGYVH
Mb1168|M.bovis_AF2122/97 AHDNPDVRAVVITGAGDKSLCAGADLKAIARRENPYHPHHGEWGIAGYRH
Mflv_2117|M.gilvum_PYR-GCK AQEDPDVRAVILTGAGDKSFCAGADLKAISRGENLFHPEHSEWGFAGYVR
Mvan_4588|M.vanbaalenii_PYR-1 AQDDPEIRAVILTGAGDKSFCAGADLKAISRGENLFHPEHTEWGFAGYVR
MSMEG_5198|M.smegmatis_MC2_155 AQNDPEVRAVVLTGAGDKSFCAGADLKAISRGENLFHPEHPEYGFAGYVS
TH_4237|M.thermoresistible__bu ---------VVLTGSGDKSFCAGADLKAIARGENLFHPDHPEWGFAGYVR
MAV_2597|M.avium_104 AQRDPEVWAVVITGAGDKSFCAGADLKAISRGENLYHAEHPEWGFAGYVH
MLBr_02402|M.leprae_Br4923 LDIDPDVGAILITGSPK-VFAAGADIKEMASLTFTDAFDADFFSAWG-KL
::**. :.** *: : *
Rv0222|M.tuberculosis_H37Rv RPPTKPLIAAVEGYALAGGTELALAADLIVAARDSAFGIPEVKRGLVAGG
MAB_4588c|M.abscessus_ATCC_199 VPPRKPIIAAVEGFALAGGTELVLSCDLVVAGRSAKFGIPEVKRGLVAGA
MMAR_4317|M.marinum_M HFIDKPTIAAVNGTALGGGTELALASDLVVADERAKFGLPEVKRGLIAAA
MUL_0972|M.ulcerans_Agy99 HFIDKPTIAAVNGTALCGGTELALASDLVVADERAKFGLPEAKRGFIAAA
Mb1168|M.bovis_AF2122/97 HFIDKPTSAAVSGTALDDGAEPALASDLVVADEHT---------------
Mflv_2117|M.gilvum_PYR-GCK HFIDKPTIAAVNGTALGGGTELALASDLVVAEERAKFGLPEVKRGLIAGA
Mvan_4588|M.vanbaalenii_PYR-1 HFIDKPTIAAVNGTALGGGTELALASDLVVAEERAKFGLPEVKRGLIAGA
MSMEG_5198|M.smegmatis_MC2_155 HFIDKPTIAAVNGTALGGGTELALASDLVVAEERAMFGLPEVKRGLIAGA
TH_4237|M.thermoresistible__bu HFMDKPTIAAVNGTALGGGTELALASDLVVARESAVFGLPEVKRGLIAGA
MAV_2597|M.avium_104 HFIDKPTIAAVNGTALGGGSELALASDLVIACESASFGLPEVKRGLIAGA
MLBr_02402|M.leprae_Br4923 AAVRTPMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGM
.* *** * ** .* * .: .**::* :
Rv0222|M.tuberculosis_H37Rv GGLLRLPERIPYAIAMELALTGDNLPAERAHELGLVNVLAEPGTALDAAI
MAB_4588c|M.abscessus_ATCC_199 GGLLRLPNRIPYQVAMELALTGESFTAEDAAKYGFINRLVDDGQALDTAL
MMAR_4317|M.marinum_M GGVFRIVNQLPRKVAMEMLLTGEPITASDAYEWGLINQVVKQGAVLDAAL
MUL_0972|M.ulcerans_Agy99 GGVFRIVNQLPRKVAMEMLLTGEPITASDAYEWGLINQVVKQGAVLDAAL
Mb1168|M.bovis_AF2122/97 --------------------------------------------------
Mflv_2117|M.gilvum_PYR-GCK GGVFRIVEQLPRKVALELIYTGEPISSADALRWGLINQVVPDGTVVDAAL
Mvan_4588|M.vanbaalenii_PYR-1 GGVFRIVEQLPRKVALELIYTGEPISSADALRWGLINQVVPDGTVVDAAL
MSMEG_5198|M.smegmatis_MC2_155 GGVFRIIEQLPRKVALELVYTGDPISAADALKWGLINQVVPDGTVVDAAL
TH_4237|M.thermoresistible__bu GGVFRIVEQLPRKVALELLFTGEPITAAEALRWGLINEVVPDGAELDAAL
MAV_2597|M.avium_104 GGVFRIVEQLPRKVALELVLTGEPMTASDALRWGLINEVVPDGTVVEAAL
MLBr_02402|M.leprae_Br4923 GGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAK
Rv0222|M.tuberculosis_H37Rv ALAEKITANGPLAVVATKRIITESRG---WSPDTMFAEQMKILVPVFTSN
MAB_4588c|M.abscessus_ATCC_199 ELAAKITANGPLAVAATKRIIIESAS---WAPEEAFAKQGEILMPIFVSE
MMAR_4317|M.marinum_M ALAARVTVNAPLSVRASKRIAYGVDDGVVADEERGWERTMREMRSLIRSE
MUL_0972|M.ulcerans_Agy99 ALAARVTVNAPLSVRASKRIAYGVDDGVVADEERGWERTMREMRSLIRSE
Mb1168|M.bovis_AF2122/97 --------------------------------------------------
Mflv_2117|M.gilvum_PYR-GCK ALAERITCNAPLAVRASKRVSYGAIDGVVGSDEPFWKQTFSEFSTLLKTE
Mvan_4588|M.vanbaalenii_PYR-1 ALAERITCNAPLAVQASKRVSYGAVDGVVGSEEPFWKQTFGEFSTLLKTE
MSMEG_5198|M.smegmatis_MC2_155 ALAERITCNAPLAVQASKRVAYGAEEGTVPAEQPSWKRTNREFVTLLKTE
TH_4237|M.thermoresistible__bu ALAERITVNAPLAVQASKRVAYGAENGTVPGDENGWKLTNREFSTLLKTE
MAV_2597|M.avium_104 ALAERITCNAPLSVQASKRVAYGADDGIIGAEEPKWERTIREFTELLKSE
MLBr_02402|M.leprae_Br4923 AVATTISQMSRSATRMAKEAVNRSFES---TLAEGLLHERRLFHSTFVTD
Rv0222|M.tuberculosis_H37Rv DAKEGAIAFAERRRPRWTGT
MAB_4588c|M.abscessus_ATCC_199 DAKEGAKAFAEKRAPVWQGK
MMAR_4317|M.marinum_M DAKEGPLAFAQKREPVWKAR
MUL_0972|M.ulcerans_Agy99 DAKEGPLAFAEKREPVWKTR
Mb1168|M.bovis_AF2122/97 --------------------
Mflv_2117|M.gilvum_PYR-GCK DAMEGPLAFAQKREPVWKAK
Mvan_4588|M.vanbaalenii_PYR-1 DAMEGPLAFAQKREPVWKAK
MSMEG_5198|M.smegmatis_MC2_155 DAMEGPLAFAQKRQPVWKAK
TH_4237|M.thermoresistible__bu DAKEGPLAFAEKREPVWKAR
MAV_2597|M.avium_104 DAKEGPLAFAEKRQPVWKAR
MLBr_02402|M.leprae_Br4923 DQSEGMAAFIEKRAPQFTHR