For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSLLTALRLNMTNIADPAADHAQRYRAALDMAEYADHHGFTAVSAEEHHLAATGWLPSPLILAAAVAGRT RNVRISINALIVPLYDPVRPAEDIAVLDNLSQGRFSFVAGMGYRPQEYHAAGKDWSRRGALMDHCLSVLL QAWGDDPFEYDGRLINVTPKPRTHPHPILFVGGMSVAAARRAARFGLPFSPPMAMPELEAVYHQELARRN KRGFVYHPENGSTVTLLHADPDEAWSRFGGFIMNEATEYSGWKRADVPRPNEAPAASIAELRSLDNVQIL TPDQLVGQLQRGRKEVVMNPLIGGLPVEVGWSSLRLLAEEVLPRVADIPDSATTKP
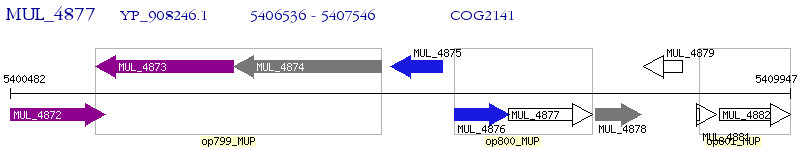
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4877 | - | - | 100% (336) | hypothetical protein MUL_4877 |
| M. ulcerans Agy99 | MUL_1265 | - | 8e-45 | 35.83% (307) | hypothetical protein MUL_1265 |
| M. ulcerans Agy99 | MUL_1043 | - | 3e-20 | 32.04% (181) | hypothetical protein MUL_1043 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3106c | - | 4e-16 | 29.56% (159) | hypothetical protein Mb3106c |
| M. gilvum PYR-GCK | Mflv_4097 | - | 7e-21 | 31.32% (182) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3079c | - | 4e-16 | 29.56% (159) | hypothetical protein Rv3079c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0366 | - | 3e-21 | 34.04% (188) | hypothetical protein MAB_0366 |
| M. marinum M | MMAR_0236 | - | 0.0 | 99.11% (336) | hypothetical protein MMAR_0236 |
| M. avium 104 | MAV_0338 | - | 5e-59 | 37.96% (324) | hypothetical protein MAV_0338 |
| M. smegmatis MC2 155 | MSMEG_4820 | - | 5e-18 | 31.46% (178) | hypothetical protein MSMEG_4820 |
| M. thermoresistible (build 8) | TH_3375 | - | 2e-32 | 31.21% (330) | PUTATIVE putative conserved hypothetical protein |
| M. vanbaalenii PYR-1 | Mvan_2248 | - | 7e-20 | 29.90% (194) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4097|M.gilvum_PYR-GCK MQFAITHPMHSHPYNPELVTGAGIARVAVAAEKAGFGGFGFTDH---PAP
Mvan_2248|M.vanbaalenii_PYR-1 MQFAITHPMHSHPYNPELVTGTGIAQVAQAAEKAGFGGFGFTDH---PAP
MSMEG_4820|M.smegmatis_MC2_155 MRYSITHPMHSHPYHPDLVSGSGVVAVAAAAEAAGFDGFGFTDH---PAP
Mb3106c|M.bovis_AF2122/97 MQFGVLTFVT--------DEGIGPAELGAALEHRGFESLFLAEHTHIPVN
Rv3079c|M.tuberculosis_H37Rv MQFGVLTFVT--------DEGIGPAELGAALEHRGFESLFLAEHTHIPVN
MUL_4877|M.ulcerans_Agy99 --MSLLTALRLNMTNIADPAADHAQRYRAALDMAEYADHHGFTA---VSA
MMAR_0236|M.marinum_M --MSLLTALRLNMTNIADPAADHAQRYRAALDMAEYADHHGFTA---VSA
MAV_0338|M.avium_104 ---MLLTVLRFNFASPHGDPRTQSRLMSAALELAQWGESHGITS---VSV
MAB_0366|M.abscessus_ATCC_1997 MKIGVVAPVELG----VTAEPDKILEFARAAERLGFSEISVVEHAVVIGD
TH_3375|M.thermoresistible__bu VGLGALTAQTRS-----GTAAESTEALTMLLDTCVLAEDEGLDS---AWV
:
Mflv_4097|M.gilvum_PYR-GCK TQR---WLEAGG------HDALDPFVAMGFAAAHTTTLRLIPNIVVLPYR
Mvan_2248|M.vanbaalenii_PYR-1 TQR---WLEAGG------HDALDPFVAMGFAAAHTSTLRLIPNIVVLPYR
MSMEG_4820|M.smegmatis_MC2_155 SQR---WLDAGG------HDAVDPFVAMAYAAAQTTTLRLVPNIVVLPYR
Mb3106c|M.bovis_AF2122/97 TQS---PYPGGGPIPEKYYRTLDPFVALAAAAATTQSLVLGTGIALIPER
Rv3079c|M.tuberculosis_H37Rv TQS---PYPGGGPIPEKYYRTLDPFVALAAAAATTQSLVLGTGIALIPER
MUL_4877|M.ulcerans_Agy99 EEH---HLAATG-------WLPSPLILAAAVAGRTRNVRISINALIVPLY
MMAR_0236|M.marinum_M EEH---HLAATG-------WLPSPLILAAAVAGRTRNVRISINALIVPLY
MAV_0338|M.avium_104 DEH---HATGHG-------WSCNPIMAAAMFLARTSTLIASVDCALGPLW
MAB_0366|M.abscessus_ATCC_1997 TQSTYPYSPTGQSHLPDDIDIPDPLELLSFVAAATTTLGLSTGVLVLPDH
TH_3375|M.thermoresistible__bu AEH---HFAADG-------FLSSPMTVLAALSQRTSRIRLSTNVMIGPLY
: .*: . * : . : *
Mflv_4097|M.gilvum_PYR-GCK NPFVVAKAGATLDLISDGRFTLAVGVGYLKREFAALGVEFDERAAVFEEA
Mvan_2248|M.vanbaalenii_PYR-1 NPFVVAKAGATLDLISEGRFTLAVGVGYLKREFTALGVDFDERAELFEES
MSMEG_4820|M.smegmatis_MC2_155 NPFVVAKSGATLDLLSGGRFTLAVGVGYLKREFAALGVAFDERTELFDES
Mb3106c|M.bovis_AF2122/97 DPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVIDER
Rv3079c|M.tuberculosis_H37Rv DPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVIDER
MUL_4877|M.ulcerans_Agy99 DPVRPAEDIAVLDNLSQGRFSFVAGMGYRPQEYHAAGKDWSRRGALMDHC
MMAR_0236|M.marinum_M DPVRLAEDIAVLDNLSQGRFSFVAGMGYRPQEYHAAGKDWSRRGALMDHC
MAV_0338|M.avium_104 NPVRLAEDIALVDNMSRGRLHTTVGLGYRPVEYDALGADFGRRGALMDSL
MAB_0366|M.abscessus_ATCC_1997 HPVVLAKRLATLDRLSRGRLRICAGVGWMREEIEACGGDFDRRGRAADEA
TH_3375|M.thermoresistible__bu DPIRLAEDAAVVDQLSRGRLMLGLGLGYRDVEFAGLGRRRQERGRRLDDL
.*. *: * :* :* **: *:*: * * * :
Mflv_4097|M.gilvum_PYR-GCK LDVITQIWTTDD-LSYDGRNFSAKGITAHPKPLTTPHPPIWVGGNTSAAR
Mvan_2248|M.vanbaalenii_PYR-1 LQVIRDAWTTDD-ITFEGKHFSAKGITAHPRPVSTPHPPIWVGGNTSAAR
MSMEG_4820|M.smegmatis_MC2_155 LEVIRGIWTTDD-FTYEGMHFSARGITAHPRPVNP--PPIWIGGNTAAAR
Mb3106c|M.bovis_AF2122/97 LRAIIEIWTQEQ-AEFHGTYVDFDPIYCWPKPVTKPYPPLYVGG-GPANF
Rv3079c|M.tuberculosis_H37Rv LRAIIEIWTQEQ-AEFHGTYVDFDPIYCWPKPVTKPYPPLYVGG-GPANF
MUL_4877|M.ulcerans_Agy99 LSVLLQAWGDDP-FEYDGRLINVT-----PKPRTHPHPILFVGGMSVAAA
MMAR_0236|M.marinum_M LSVLLQAWGDDP-FEYDGRLINVT-----PKPRTHPHPILFVGGMSVAAA
MAV_0338|M.avium_104 LERMLAVWSGG------GAPAGIC-----TGTWTRPHPPLYVGGGARATA
MAB_0366|M.abscessus_ATCC_1997 IAVMRALWSTDGPTSHDGEFYSFRNAYSYPKPVRPQGVPLYIGGHSKAAA
TH_3375|M.thermoresistible__bu INVLRQAWSGKP--VEHHGLGTYKTPPVTPLPFQQDGPPVLVGAFAEAGL
: : * . . : :*. *
Mflv_4097|M.gilvum_PYR-GCK ARVAKYAQGWCPFRAPAMLAQTARTAALE--------TTEHLAKGIDDLR
Mvan_2248|M.vanbaalenii_PYR-1 ARVARYAQGWCPFRAPAVLAQTARTAALE--------TVEHLKAGIDDLR
MSMEG_4820|M.smegmatis_MC2_155 ARVARHGDGWCPFAAPPGLAQTAGTAAID--------SIERLAAGIADLR
Mb3106c|M.bovis_AF2122/97 PRIARLNAGWIAISP----------------------SPQRLSGPLQRLR
Rv3079c|M.tuberculosis_H37Rv PRIARLNAGWIAISP----------------------SPQRLSGPLQRLR
MUL_4877|M.ulcerans_Agy99 RRAARFGLPFSPPMAMPELEAVYHQELARRNKRGFVYHP-ENGSTVTLLH
MMAR_0236|M.marinum_M RRAAHFGLPFSPPMAMPELEAVYHQELARRNKRGFVYHP-ENGSTVTLLH
MAV_0338|M.avium_104 RRAARFRLPLSLADHLPEIADYYRELCAEIDLAPVVLMPGPLNRGMIYLH
MAB_0366|M.abscessus_ATCC_1997 RRAGRLGSGFQPLGLVG----------------------DDLTAAVGVMR
TH_3375|M.thermoresistible__bu RRAVRLGNGWLAPELRHWRGLEKRVAALGLDSR---TDPFHVAVTVNAFV
* : : :
Mflv_4097|M.gilvum_PYR-GCK RRLEDAGRDPADVDITFTNDAGGAPGTDD---------------------
Mvan_2248|M.vanbaalenii_PYR-1 RRLEGTGRDPFDIDITFTNDAGGSPGAAD---------------------
MSMEG_4820|M.smegmatis_MC2_155 AKLDAAQRDPSTVDIVFTNFDGGNPGDDD---------------------
Mb3106c|M.bovis_AF2122/97 AMAGG--------DVPVTVCQWGEAAAKD---------------------
Rv3079c|M.tuberculosis_H37Rv AMAGG--------DVPVTVCQWGEAAAKD---------------------
MUL_4877|M.ulcerans_Agy99 ADPDEAWSRFGGFIMNEATEYSGWKRADVPRPNEAP--AASIAELRSLDN
MMAR_0236|M.marinum_M ADPDEAWSRFGGFIMNEATEYSGWKRADVPRPNEAP--AASIAELRGLDN
MAV_0338|M.avium_104 EDPERAWAELGDHILWEAVTYGGWSTDQRSLMHLPG--VQTLDEVRASGR
MAB_0366|M.abscessus_ATCC_1997 AAAVDAGRDPRGLDLVLGHGLHRVDQAAL---------------------
TH_3375|M.thermoresistible__bu AARDAWETVRPGVAHVAGQYRSWLVESGDLPALEGKEFEYEADETGKPPQ
Mflv_4097|M.gilvum_PYR-GCK ---FNADEFLAGAE---------ELERAGVTWLQVNVPGDSLAHAVEAIE
Mvan_2248|M.vanbaalenii_PYR-1 ---FNADEFLSGAA---------ELERAGVTWIQVTVPGDSLAHTLEAIE
MSMEG_4820|M.smegmatis_MC2_155 ---FDADAYLAGVD---------QLAALGVTWLHVTIPGDSLAHALETIE
Mb3106c|M.bovis_AF2122/97 ---LEGYRHLG------------------VERVLLELPTEPRDPTLRYLD
Rv3079c|M.tuberculosis_H37Rv ---LEGYRHLG------------------VERVLLELPTEPRDPTLRYLD
MUL_4877|M.ulcerans_Agy99 VQILTPDQLVGQLQ---------RGRKE--VVMNPLIGGLPVEVGWSSLR
MMAR_0236|M.marinum_M VQILTPDQLVGQLQ---------RGRKE--VVMNPLIGGLPVEVGWSSLR
MAV_0338|M.avium_104 YRFLTPDQLIAEVR---------DAPDFGPIVLHPLVGGMPVEEAWQSVQ
MAB_0366|M.abscessus_ATCC_1997 ----DQAAHAG----------------AGRMLLSASRRATTLEAVLEEMA
TH_3375|M.thermoresistible__bu FVAGTPEQCVAQLRPWWRILARLPEHVQPHLILGMTFPGVSREATFESVR
. : . :
Mflv_4097|M.gilvum_PYR-GCK GFGETVIAQR-----------
Mvan_2248|M.vanbaalenii_PYR-1 EFGGSVIGRP-----------
MSMEG_4820|M.smegmatis_MC2_155 GFGKSVIAR------------
Mb3106c|M.bovis_AF2122/97 KLQAELARLA-----------
Rv3079c|M.tuberculosis_H37Rv KLQAELARLA-----------
MUL_4877|M.ulcerans_Agy99 LLAEEVLPRVADIPDSATTKP
MMAR_0236|M.marinum_M LLAEEVLPRVADIPDSATTKP
MAV_0338|M.avium_104 LLTDKVLPALKG---------
MAB_0366|M.abscessus_ATCC_1997 GCATRLELGGPR---------
TH_3375|M.thermoresistible__bu LLAREVVPALNAEE-------
: