For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RYSITHPMHSHPYHPDLVSGSGVVAVAAAAEAAGFDGFGFTDHPAPSQRWLDAGGHDAVDPFVAMAYAAA QTTTLRLVPNIVVLPYRNPFVVAKSGATLDLLSGGRFTLAVGVGYLKREFAALGVAFDERTELFDESLEV IRGIWTTDDFTYEGMHFSARGITAHPRPVNPPPIWIGGNTAAARARVARHGDGWCPFAAPPGLAQTAGTA AIDSIERLAAGIADLRAKLDAAQRDPSTVDIVFTNFDGGNPGDDDFDADAYLAGVDQLAALGVTWLHVTI PGDSLAHALETIEGFGKSVIAR*
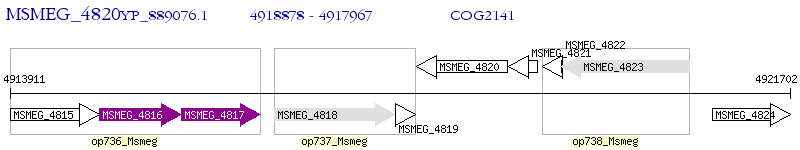
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4820 | - | - | 100% (303) | hypothetical protein MSMEG_4820 |
| M. smegmatis MC2 155 | MSMEG_0654 | - | 9e-30 | 38.17% (186) | hypothetical protein MSMEG_0654 |
| M. smegmatis MC2 155 | MSMEG_2811 | - | 2e-29 | 40.00% (215) | hydride transferase 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3106c | - | 3e-25 | 37.31% (193) | hypothetical protein Mb3106c |
| M. gilvum PYR-GCK | Mflv_4097 | - | 1e-129 | 70.16% (305) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3079c | - | 3e-25 | 37.31% (193) | hypothetical protein Rv3079c |
| M. leprae Br4923 | MLBr_00348 | - | 2e-12 | 29.69% (192) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0366 | - | 8e-28 | 34.04% (235) | hypothetical protein MAB_0366 |
| M. marinum M | MMAR_0393 | - | 1e-135 | 75.91% (303) | hypothetical protein MMAR_0393 |
| M. avium 104 | MAV_5129 | - | 1e-135 | 74.59% (303) | hypothetical protein MAV_5129 |
| M. thermoresistible (build 8) | TH_2146 | - | 1e-126 | 70.82% (305) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1043 | - | 1e-132 | 76.61% (295) | hypothetical protein MUL_1043 |
| M. vanbaalenii PYR-1 | Mvan_2248 | - | 1e-129 | 70.82% (305) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3106c|M.bovis_AF2122/97 ---------MQFGVLTFVTDEGIGPAELGAALEHRGFESLFLAEHTHIPV
Rv3079c|M.tuberculosis_H37Rv ---------MQFGVLTFVTDEGIGPAELGAALEHRGFESLFLAEHTHIPV
MMAR_0393|M.marinum_M -MRFIITHPMHSHPYHPELASGAGVAAVAAAAERAGFHGFGFTDH---PA
MUL_1043|M.ulcerans_Agy99 ---------MHSHPYHPELASGAGVAAVAAAAERAGFHGFGFTDH---PA
MAV_5129|M.avium_104 -MRFTITHPMHSHPYNRELVSGDAIAKVAAATEAAGIHGFGFTDH---PA
MSMEG_4820|M.smegmatis_MC2_155 -MRYSITHPMHSHPYHPDLVSGSGVVAVAAAAEAAGFDGFGFTDH---PA
Mflv_4097|M.gilvum_PYR-GCK -MQFAITHPMHSHPYNPELVTGAGIARVAVAAEKAGFGGFGFTDH---PA
Mvan_2248|M.vanbaalenii_PYR-1 -MQFAITHPMHSHPYNPELVTGTGIAQVAQAAEKAGFGGFGFTDH---PA
TH_2146|M.thermoresistible__bu -MRFAFTYPMDSHPYHPDLATGAAVARVAATAETAGFSGFGFTDH---PA
MAB_0366|M.abscessus_ATCC_1997 -MKIGVVAPVELG----VTAEPDKILEFARAAERLGFSEISVVEHAVVIG
MLBr_00348|M.leprae_Br4923 MVERANAMKLGLQLGYWAAQPPGNAAELVAAAEGAGFDAVFTGEAWG---
: . : * *: . :
Mb3106c|M.bovis_AF2122/97 NTQS---PYPGGGPIPEKYYRTLDPFVALAAAAATTQSLVLGTGIALIPE
Rv3079c|M.tuberculosis_H37Rv NTQS---PYPGGGPIPEKYYRTLDPFVALAAAAATTQSLVLGTGIALIPE
MMAR_0393|M.marinum_M PSQR---WLEAGG------HDSLDPFVALGFAAAQTTSLRLIPNIVVLPY
MUL_1043|M.ulcerans_Agy99 PSQR---WLEAGG------HDSLDPFVALGFAAAQTTSLRLIPNIVVLPY
MAV_5129|M.avium_104 PSQR---WLEAGG------HDALDPFVALGFAAATTSTLRLIPNIVVLPY
MSMEG_4820|M.smegmatis_MC2_155 PSQR---WLDAGG------HDAVDPFVAMAYAAAQTTTLRLVPNIVVLPY
Mflv_4097|M.gilvum_PYR-GCK PTQR---WLEAGG------HDALDPFVAMGFAAAHTTTLRLIPNIVVLPY
Mvan_2248|M.vanbaalenii_PYR-1 PTQR---WLEAGG------HDALDPFVAMGFAAAHTSTLRLIPNIVVLPY
TH_2146|M.thermoresistible__bu PTQK---WLDHGG------HDTLDPFVAMGFAAAHTTTLRLIPHIVVLPY
MAB_0366|M.abscessus_ATCC_1997 DTQSTYPYSPTGQSHLPDDIDIPDPLELLSFVAAATTTLGLSTGVLVLPD
MLBr_00348|M.leprae_Br4923 ----------------------SDAYTPLAWWGSSTQRVRLGTSVVQLSA
*. :. .: * : * . : :.
Mb3106c|M.bovis_AF2122/97 RDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVIDE
Rv3079c|M.tuberculosis_H37Rv RDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVIDE
MMAR_0393|M.marinum_M RNPFVVAKSAATLDLLSGGRFILAVGVGYLKREFAALGVDHAERAELFEE
MUL_1043|M.ulcerans_Agy99 RNPFVVAKSAATLDLLSGGRFILAVGVGYLKREFAALGVDHAERAELFDE
MAV_5129|M.avium_104 RNPFVVAKSGATLDLLSGGRFTLAVGVGYLKREFAALGVSYDERAELFEE
MSMEG_4820|M.smegmatis_MC2_155 RNPFVVAKSGATLDLLSGGRFTLAVGVGYLKREFAALGVAFDERTELFDE
Mflv_4097|M.gilvum_PYR-GCK RNPFVVAKAGATLDLISDGRFTLAVGVGYLKREFAALGVEFDERAAVFEE
Mvan_2248|M.vanbaalenii_PYR-1 RNPFVVAKAGATLDLISEGRFTLAVGVGYLKREFTALGVDFDERAELFEE
TH_2146|M.thermoresistible__bu RNPFVVAKAGASVDLLSGGRFTLGVGVGYLKREFAALGVDFEERAALFEE
MAB_0366|M.abscessus_ATCC_1997 HHPVVLAKRLATLDRLSRGRLRICAGVGWMREEIEACGGDFDRRGRAADE
MLBr_00348|M.leprae_Br4923 RTPTACAMAALTLDHLSGGRHILGLGVSGPQVVEGWYGQPFPKPLSRTRE
: * . * ::* :* ** : **. : * *
Mb3106c|M.bovis_AF2122/97 RLRAIIEIWTQE-QAEFHGTYV-------DFDPIYCWPKPVTKPY---PP
Rv3079c|M.tuberculosis_H37Rv RLRAIIEIWTQE-QAEFHGTYV-------DFDPIYCWPKPVTKPY---PP
MMAR_0393|M.marinum_M ALQVIRAVWTSD-DISFEGRHF-------SARGITAHPRPASNPY---PP
MUL_1043|M.ulcerans_Agy99 ALQVIRAVWTSD-DISFEGRHF-------SARGITAHPRPASNPY---PP
MAV_5129|M.avium_104 ALQVIRAVWTGD-DISFEGRHF-------SARGITAHPRPVSSPH---PP
MSMEG_4820|M.smegmatis_MC2_155 SLEVIRGIWTTD-DFTYEGMHF-------SARGITAHPRPVNP-----PP
Mflv_4097|M.gilvum_PYR-GCK ALDVITQIWTTD-DLSYDGRNF-------SAKGITAHPKPLTTPH---PP
Mvan_2248|M.vanbaalenii_PYR-1 SLQVIRDAWTTD-DITFEGKHF-------SAKGITAHPRPVSTPH---PP
TH_2146|M.thermoresistible__bu ALEVIRAIWTTD-NVSYTGRHF-------TADGITAHPHPVTKPH---PP
MAB_0366|M.abscessus_ATCC_1997 AIAVMRALWSTDGPTSHDGEFY-------SFRNAYSYPKPVRPQG---VP
MLBr_00348|M.leprae_Br4923 YIDIVRQVWAREAPVVSDGQHYPLPLTGAGATGLGKALKPITHPLRADIP
: : *: : * :* *
Mb3106c|M.bovis_AF2122/97 LYVGG-GPANFPRIARLNAGWIAISP-----------------------S
Rv3079c|M.tuberculosis_H37Rv LYVGG-GPANFPRIARLNAGWIAISP-----------------------S
MMAR_0393|M.marinum_M IWVGGNTAAARRRVAAHGDGWCPFPAPAPLAQTA---------GTAAIDS
MUL_1043|M.ulcerans_Agy99 IWVGGNTAAARRRVAVHGDGWCPFPAPAPLAQTA---------GTAAIDS
MAV_5129|M.avium_104 IWIGGNTAAARQRVADYGDGWCPFPAPPQLARTA---------GTAVIDS
MSMEG_4820|M.smegmatis_MC2_155 IWIGGNTAAARARVARHGDGWCPFAAPPGLAQTA---------GTAAIDS
Mflv_4097|M.gilvum_PYR-GCK IWVGGNTSAARARVAKYAQGWCPFRAPAMLAQTA---------RTAALET
Mvan_2248|M.vanbaalenii_PYR-1 IWVGGNTSAARARVARYAQGWCPFRAPAVLAQTA---------RTAALET
TH_2146|M.thermoresistible__bu IWIGGNTGAARQRVADHGAGWCPFPAPAVLAQTA---------RTAAMDS
MAB_0366|M.abscessus_ATCC_1997 LYIGGHSKAAARRAGRLGSGFQPLGLVG----------------------
MLBr_00348|M.leprae_Br4923 IMLGAEGPKNVALAAEICDGWLPIFFSPRMADMYNEWLDEGFARTGARRS
: :*. . *: .:
Mb3106c|M.bovis_AF2122/97 PQRLSGPLQRLRAMAGG--------DVPVTVCQWGEAAA-KDLEGYRHLG
Rv3079c|M.tuberculosis_H37Rv PQRLSGPLQRLRAMAGG--------DVPVTVCQWGEAAA-KDLEGYRHLG
MMAR_0393|M.marinum_M ADRLAAGIDDLRRKCDAAGRDFSAIDVTFTNFDGGGPAS-ADFNADAYLG
MUL_1043|M.ulcerans_Agy99 ADRLAAGIDDLRRKCDAAGRDFSAIDVTFTNFDGGGPAS-ADFNADAYLG
MAV_5129|M.avium_104 VAKLADGIADLRRRCDAAQRDWSTIDITFTNFEGGSPAD-DEFNADAYLD
MSMEG_4820|M.smegmatis_MC2_155 IERLAAGIADLRAKLDAAQRDPSTVDIVFTNFDGGNPGD-DDFDADAYLA
Mflv_4097|M.gilvum_PYR-GCK TEHLAKGIDDLRRRLEDAGRDPADVDITFTNDAGGAPGT-DDFNADEFLA
Mvan_2248|M.vanbaalenii_PYR-1 VEHLKAGIDDLRRRLEGTGRDPFDIDITFTNDAGGSPGA-ADFNADEFLS
TH_2146|M.thermoresistible__bu AEALAAGITDLHRRLEATGRDPADVDVVFSNLTGGTPGR-ADFNADAYLD
MAB_0366|M.abscessus_ATCC_1997 -DDLTAAVGVMRAAAVDAGRDPRGLDLVLG-HGLHRVDQ-AALDQAAHAG
MLBr_00348|M.leprae_Br4923 REDFEICASAQIVVTEDRAAAFAGIKPFLALYMGGMGAEGTNFHADVYRR
: . . :. .
Mb3106c|M.bovis_AF2122/97 ---------VERVLLELPTEPRDPTLRYLDKLQAELARLA----------
Rv3079c|M.tuberculosis_H37Rv ---------VERVLLELPTEPRDPTLRYLDKLQAELARLA----------
MMAR_0393|M.marinum_M GLEKLATLGVTWVHVGLPGDGLAHALECIDRFRVQVIEAA----------
MUL_1043|M.ulcerans_Agy99 GLEKLAMLGVTWVHVGLPGDGLAHALECIDRFRVQVIEAA----------
MAV_5129|M.avium_104 GLDKLAKLGVTWVSVHLPGDSMAHALETLDRFRTLVVDAA----------
MSMEG_4820|M.smegmatis_MC2_155 GVDQLAALGVTWLHVTIPGDSLAHALETIEGFGKSVIAR-----------
Mflv_4097|M.gilvum_PYR-GCK GAEELERAGVTWLQVNVPGDSLAHAVEAIEGFGETVIAQR----------
Mvan_2248|M.vanbaalenii_PYR-1 GAAELERAGVTWIQVTVPGDSLAHTLEAIEEFGGSVIGRP----------
TH_2146|M.thermoresistible__bu GVAKLGALGVTWLQVPIPGDSLNHALEAIEAFGSEVIGKL----------
MAB_0366|M.abscessus_ATCC_1997 -------AGRMLLSASRRATTLEAVLEEMAGCATRLELGGPR--------
MLBr_00348|M.leprae_Br4923 MGYAEVVDEVTALFRSNQKDKAAEIIPNELVDSTMIVGDVDYVCKQIAAW
: : :
Mb3106c|M.bovis_AF2122/97 -------------------------
Rv3079c|M.tuberculosis_H37Rv -------------------------
MMAR_0393|M.marinum_M -------------------------
MUL_1043|M.ulcerans_Agy99 -------------------------
MAV_5129|M.avium_104 -------------------------
MSMEG_4820|M.smegmatis_MC2_155 -------------------------
Mflv_4097|M.gilvum_PYR-GCK -------------------------
Mvan_2248|M.vanbaalenii_PYR-1 -------------------------
TH_2146|M.thermoresistible__bu -------------------------
MAB_0366|M.abscessus_ATCC_1997 -------------------------
MLBr_00348|M.leprae_Br4923 SAAGVTMMMVSASSVEQVRDLAGLF