For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IGLGGVWVLDGLEVTMVGNVSARLTEHGSGINLTPAQIGVAASFYIAGACLGALFFGHLTDRFGRRNLFL LTLAVYLTATVATAFAFAPWYFFIARFFTGSGIGGEYAAINSAIDELIPARVRGRVDLIINGTYWLGSAA GAAGALVLLDTSNFAPNMGWRLAFGLGAIFGIFVLLVRRNVPESPRWLFIHGREQEAEQIVREIEEEVER DTGQRLPDPPGRPLKIRQRQRISLWEIARVAFTRYPRRAFLGLALFIGQAFLYNGVTFNLGTLLSGFYGV ESGHVPLFFIVWALSNFVGPLVLGRLFDTVGRKPMITMSYIGSAVIAVVLAVIFVNQTGGVWTFIIVLAA AFFLASAGASAAYLTVSEIFPMETRALAIAFFYAVGTAIGGISGPLLFGQLIESGQRSQVFWSFIIGAAV MAVAGLVELWLGIAAEQRPLEELALPLTVVDAEDDDEPADGAGARGAQHPGS*
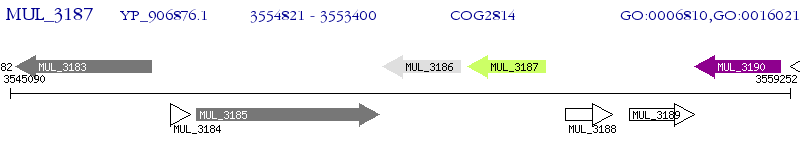
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3187 | - | - | 100% (473) | sugar transporter |
| M. ulcerans Agy99 | MUL_0949 | - | 1e-16 | 27.02% (396) | integral membrane transport protein |
| M. ulcerans Agy99 | MUL_1444 | sugI | 3e-14 | 22.54% (426) | sugar-transport integral membrane protein SugI |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3503c | kgtP | 4e-17 | 23.64% (368) | dicarboxylic acid transport integral membrane protein KgtP |
| M. gilvum PYR-GCK | Mflv_3055 | - | 7e-20 | 24.76% (416) | sugar transporter |
| M. tuberculosis H37Rv | Rv3476c | kgtP | 2e-16 | 23.37% (368) | dicarboxylic acid transport integral membrane protein KgtP |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0365c | - | 2e-19 | 25.12% (430) | sugar-transport integral membrane protein |
| M. marinum M | MMAR_2574 | - | 0.0 | 99.79% (473) | sugar transporter |
| M. avium 104 | MAV_3053 | - | 0.0 | 81.43% (463) | sugar transport protein |
| M. smegmatis MC2 155 | MSMEG_3536 | - | 0.0 | 72.71% (469) | sugar transport protein |
| M. thermoresistible (build 8) | TH_3400 | - | 1e-16 | 24.16% (447) | PUTATIVE General substrate transporter |
| M. vanbaalenii PYR-1 | Mvan_3472 | - | 1e-22 | 25.74% (408) | sugar transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3187|M.ulcerans_Agy99 ------------------------------MIGLGG--VWVLDGLEVTMV
MMAR_2574|M.marinum_M MSARAQTGTISTHVPARLDRLPWGRFHWRVVIGLGG--VWVLDGLEVTMV
MAV_3053|M.avium_104 MSAGAQTNTITTHVPARLDRLPWSRFHWRVVIGLGG--VWVLDGLEVTMV
MSMEG_3536|M.smegmatis_MC2_155 MSSGVQTGTITTQVPSRLDRLPWSRFHWRVVLGLGG--VWILDGLEVTMV
Mb3503c|M.bovis_AF2122/97 -------MTVSIAPPSRPSQAETRRAIWNTIRGSSGNLVEWYDVYVYTVF
Rv3476c|M.tuberculosis_H37Rv -------MTVSIAPPSRPSQAETRRAIWNTIRGSSGNLVEWYDVYVYTVF
Mflv_3055|M.gilvum_PYR-GCK --MAGQGGPAGESPELYEKGQDFSSGKTAIRIASVAALGGLLFGYDSAVI
Mvan_3472|M.vanbaalenii_PYR-1 --MAGQGGPVGDGPEIFEQGQEFSSGKTAVRIASVAALGGLLFGYDSAVI
MAB_0365c|M.abscessus_ATCC_199 ---------------MSLITDLRSTSRLGVMVAVTAAAVGVIYGYDSSNI
TH_3400|M.thermoresistible__bu ---LTTERTDPPQDPSVEQEQPPGVLRKAIAASAVGNATEWFDYGLYAYG
. . :
MUL_3187|M.ulcerans_Agy99 GNVSARLTEHGSGINLTPAQIGVAASFYIAGACLGALFFGHLTDRFGRRN
MMAR_2574|M.marinum_M GNVSARLTEHGSGINLTPAQIGVAASFYIAGACLGALFFGHLTDRFGRRN
MAV_3053|M.avium_104 GNVSARLMEPGSGIALNPAQIGMAAAIYIAGACSGALFFGHLTDRFGRRN
MSMEG_3536|M.smegmatis_MC2_155 GNVASRLTESGSGIELSAGQIGIAAAVYITGACLGALFFGQLTDRFGRKK
Mb3503c|M.bovis_AF2122/97 ATYFEDQFFDRADRNSTVYVYAIFAVTFVT-RPVGSWFFGRFADRRGRRA
Rv3476c|M.tuberculosis_H37Rv ATYFEDQFFDRADRNSTVYVYAIFAVTFVT-RPVGSWFLGRFADRRGRRA
Mflv_3055|M.gilvum_PYR-GCK NGAVASIQEDFG---IGNYALGLAVASALLGAAVGALSAGRIADRIGRIA
Mvan_3472|M.vanbaalenii_PYR-1 NGAVDSIQEDFG---IGNAELGFAVASALLGAAAGAMTAGRIADRIGRIA
MAB_0365c|M.abscessus_ATCC_199 AGALLFLTEDLH---LSTHDQQVAATAVVIGEIFGALIGGPLSNKIGRKR
TH_3400|M.thermoresistible__bu VTYISVAIFPGDGATATLLALMTFAVSFLV-RPLGGFVWGPLGDRLGRRQ
. : *. * : :: **
MUL_3187|M.ulcerans_Agy99 LFLLTLAVYLTATVATAFA--------FAPWYFFIARFFTGSGIGGEYAA
MMAR_2574|M.marinum_M LFLLTLAVYLTATVATAFA--------FAPWYFFIARFFTGSGIGGEYAA
MAV_3053|M.avium_104 LFILTLALYLIATVATAFA--------FAPWYFFLTRFFTGAGIGGEYAA
MSMEG_3536|M.smegmatis_MC2_155 LFLLTLAVYLVATVATAFA--------FAPWFFFVCRFFTGAGIGGEYAA
Mb3503c|M.bovis_AF2122/97 ALTFSVSLMAACSLIVALVPSRSSIGVAAPILLILCRLVQGFATGGEYGT
Rv3476c|M.tuberculosis_H37Rv ALTFSVSLMAACSLIVALVPSRSSIGVAAPILLILCRLVQGFATGGEYGT
Mflv_3055|M.gilvum_PYR-GCK VMKIAAVLFLLSAFGTAFAP--------ETITLVIGRIVGGVGVGVASVI
Mvan_3472|M.vanbaalenii_PYR-1 VMKIAAVLFLVSAFGTGFAH--------EVWAVVLFRIVGGIGVGVASVI
MAB_0365c|M.abscessus_ATCC_199 SMVLVAATFGIFSLLSALAV--------DLNTLAAARFLLGLTVGVSVVV
TH_3400|M.thermoresistible__bu VLAITILLMAGATFCVGLVPSYDSIGLWAPVLMVILRMIQGFSTGGEYGG
: : :. .:. . *:. * *
MUL_3187|M.ulcerans_Agy99 INSAIDELIPARVRGRVDLIINGTYWLGSAAGAAGALVLLDTSN------
MMAR_2574|M.marinum_M INSAIDELIPARVRGRVDLIINGTYWLGSAAGAAGALVLLDTSN------
MAV_3053|M.avium_104 INSAIDELIPARVRGRVDLVINGTYWLGSAAGAGGALILLDTSN------
MSMEG_3536|M.smegmatis_MC2_155 INSAIDELIPARVRGRVDLIINGSYWLGAAGGALGALLLLNTAI------
Mb3503c|M.bovis_AF2122/97 SVTYMSEAATRERRGYFSSFQYVTLVGGHVLAQFTLLVILAVFTRE----
Rv3476c|M.tuberculosis_H37Rv SATYMSEAATRERRGYFSSFQYVTLVGGHVLAQFTLLVILAVFTRE----
Mflv_3055|M.gilvum_PYR-GCK APAYIAETSPPGIRGRLGSLQQLAIVSGIFASFAVNYLLQWLAGGPNEPL
Mvan_3472|M.vanbaalenii_PYR-1 APAYIAETSPPGIRGRLGSLQQLAIVSGIFASFAVNWLLQWAAGGPNEPL
MAB_0365c|M.abscessus_ATCC_199 VPVFVAESSPTKIRGSMLVLYQLACVTGIIAGYLIAWALSSTGS------
TH_3400|M.thermoresistible__bu AATFMAEYAPSRRRGLLGSFLEFGTLAGFSLGALMMLGVSLMLSDD----
: * . ** . . * . :
MUL_3187|M.ulcerans_Agy99 FAPNMGWRLAFGLGAIFGIFVLLVRRNVPESPRWLFIHGREQEAEQIVRE
MMAR_2574|M.marinum_M FAPNMGWRLAFGLGAIFGIFVLLVRRNVPESPRWLFIHGREQEAEQIVRE
MAV_3053|M.avium_104 FAADLGWRLAFGIGAILGIFVLLVRRNVPESPRWLFIHGREEEAEHIVGE
MSMEG_3536|M.smegmatis_MC2_155 FPADIGWRIAFGIGAVFGLVVLVVRRHVPESPRWLFIHGQEDEAERVVGE
Mb3503c|M.bovis_AF2122/97 QVHEFGWRIGFAVGGGAAIVVFWLRRTMDES-----------LSQERLTA
Rv3476c|M.tuberculosis_H37Rv QVHEFGWRIGFAVGGGAAIVVFWLRRTMDES-----------LSQERLTA
Mflv_3055|M.gilvum_PYR-GCK WLGMDAWRWMFLAMAVPAVVYGGLAFTIPESPRYLVATHKIPEARRVLSM
Mvan_3472|M.vanbaalenii_PYR-1 WFGLDAWRWMFLAMALPAVLYGVLAFTIPESPRYLVASHKIPEARRVLSM
MAB_0365c|M.abscessus_ATCC_199 ------WRLMLGLAAVPAALVLIALLKLPDTARWYMMRGDRARAREVLSM
TH_3400|M.thermoresistible__bu QMNTWGWRVPFLIAAPLGLIGLYLRSRLDDTPIFRELQESDREEKSIWTE
** : . . . : :: .
MUL_3187|M.ulcerans_Agy99 IEEEVERDTGQRLPDPPGRPLKIRQRQRISLWEIARVAFTRYPRRAFLGL
MMAR_2574|M.marinum_M IEEEVERDTGQRLPDPPGRPLKIRQRQRISLWEIARVAFTRYPRRAFLGL
MAV_3053|M.avium_104 IEEAVQQQTGRPLPEPQGKALRIRQRTAISFREIAAVAFKLYPRRAVLGL
MSMEG_3536|M.smegmatis_MC2_155 IEREVERSTGRPLDEPEG-SLTVRQRKTISFVEIAKVAFTKYPKRAVLGL
Mb3503c|M.bovis_AF2122/97 IKAGRDHDSGS-----------------------LRELATHYWKPLLLCF
Rv3476c|M.tuberculosis_H37Rv IKAGRDHDSGS-----------------------LRELATHYWKPLLLCF
Mflv_3055|M.gilvum_PYR-GCK LLGQKNLEITITRIR------ETLEREDRPSWRDLKKPTGGIYGIVWVGL
Mvan_3472|M.vanbaalenii_PYR-1 LLGKKNLEITITRIR------ETLEREDKPSWRDMRKPTGGLYGIVWVGL
MAB_0365c|M.abscessus_ATCC_199 VDRDVDIDHELDEIAEALR----AEGGGSVRARLREMVTAPYRRATFFVV
TH_3400|M.thermoresistible__bu FKD----------------------------------LLVEYRGQILRLG
.
MUL_3187|M.ulcerans_Agy99 ALFIGQAFLYNGVTFNLGTLLSGFYGVESGHVPL-FFIVWALSNFVGPLV
MMAR_2574|M.marinum_M ALFIGQAFLYNGVTFNLGTLLSGFYGVESGHVPL-FFIVWALSNFVGPLV
MAV_3053|M.avium_104 ALFIGQAFLYNGVTFNLGTLLSQFYAVPSGMVPV-FFVLWALSNFAGPLL
MSMEG_3536|M.smegmatis_MC2_155 ALFIGQAFIYNAFTFNLGTLLSGFFDVASGAVPI-FYAVWALSNFAGPIL
Mb3503c|M.bovis_AF2122/97 LVTLGGTVAFYTYSVNAPAIVKSVYGSQAMTATW-INLVGLILLMMLQPI
Rv3476c|M.tuberculosis_H37Rv LVTLGGTVAFYTYSVNAPAIVKSVYGSQAMTATW-INLVGLILLMMLQPI
Mflv_3055|M.gilvum_PYR-GCK GLSIFQQFVGINVIFYYSNVLWQAVGFSADESAV-YTVITSVVNVLTTLI
Mvan_3472|M.vanbaalenii_PYR-1 GLSIFQQFVGINVIFYYSNVLWQAVGFSADESAV-YTVITSVINVLTTLI
MAB_0365c|M.abscessus_ATCC_199 GLGFFIQITGINAVVYYGPRIFEAMGMSGYFAKLGLPALVQVAALLAVFI
TH_3400|M.thermoresistible__bu GLVVALNVVNYTLLSYMPTYLQTSIGLSTDTALV-VPIIGMLSMMVFLPF
: . . : : : . .
MUL_3187|M.ulcerans_Agy99 LGRLFDTVGRKPMITMSYIGSAVIAVVLAVIFVNQTG-------------
MMAR_2574|M.marinum_M LGRLFDTVGRKPMITMSYIGSAVIAVVLAVIFVNQTG-------------
MAV_3053|M.avium_104 LGHLFDTVGRKQMITLTYIGSAVVVVALALVFLTQAG-------------
MSMEG_3536|M.smegmatis_MC2_155 LGRLFDTVGRKPMIALAYLGSAAVAVILTVLFVNEIG-------------
Mb3503c|M.bovis_AF2122/97 GGMISDKIGRKPLLLWFGVG-GLIYTYVLVTYLPETR-------------
Rv3476c|M.tuberculosis_H37Rv GGMISDKIGRKPLLLWFGVG-GLIYTYVLVTYLPETR-------------
Mflv_3055|M.gilvum_PYR-GCK AIALIDKIGRKPLLLIGSVGMAVTLSTMAFIFANAELIENEAGEMVPSLP
Mvan_3472|M.vanbaalenii_PYR-1 AIALIDKIGRKPLLLIGSSGMAVTLITMAVIFANATVGADGN----PSLP
MAB_0365c|M.abscessus_ATCC_199 SMSSIDRMGRRPILMIGIGIMIVADALLVGVFAVGGSSFG----------
TH_3400|M.thermoresistible__bu AGLLSDRFGRKPMWWISLGGLLALVVPMFMLMSTSVP-------------
* .**: : :
MUL_3187|M.ulcerans_Agy99 GVWTFIIVLAAAFFLASAGAS---AAYLTVSEIFPMETRALAIAFFYAVG
MMAR_2574|M.marinum_M GVWTFIIVLAAAFFLASAGAS---AAYLTVSEIFPMETRALAIAFFYAVG
MAV_3053|M.avium_104 GVWAFIGVLIVAFFLASAGAS---AAYLTVSEIFPMETRALAIAFFYAMG
MSMEG_3536|M.smegmatis_MC2_155 GVWLFMLVLGVCFFLASSGAS---AAYLTVSEIFPMETRALAIAFFYAIG
Mb3503c|M.bovis_AF2122/97 SPTMSFLLVAVGYVILTGYCS---INALVKSELFPAHVRALGVGVGYALA
Rv3476c|M.tuberculosis_H37Rv SPTMSFLLVAVGYVILTGYCS---INALVKSELFPAHVRALGVGVGYALA
Mflv_3055|M.gilvum_PYR-GCK GASGVIALIAANLFVVAFGMSWGPVVWVLLGEMFPNRIRAAALGLAAAGQ
Mvan_3472|M.vanbaalenii_PYR-1 GASGVIALIAANLFVVAFGMSWGPVVWVLLGEMFPNRIRAAALGLAAAGQ
MAB_0365c|M.abscessus_ATCC_199 GLLTFLGFLGIVLIAVGFTFGFGSLVWVYAGESFPARLRSYGASAMLTSD
TH_3400|M.thermoresistible__bu GAIIGFAVLGLLYVPQLATIS------ATFPAMFPTHVRYAGFAIAYNVS
. : .: . . ** . * . .
MUL_3187|M.ulcerans_Agy99 TAIGGISGPLLFGQLIESGQR-SQVFWSFIIGAAVMAVAGLVELWLGIAA
MMAR_2574|M.marinum_M TAIGGISGPLLFGQLIESGQR-SQVFWSFIIGAAVMAVAGLVELWLGIAA
MAV_3053|M.avium_104 TAIGGITGPLLFGQLIDSGQR-DHVVWSFLIGAVVMAAAGLVELWLGIAA
MSMEG_3536|M.smegmatis_MC2_155 TAVGGITGPLLFGQLIESGER-GLVAVSFLIGAAVMAVGGVVELILGVKA
Mb3503c|M.bovis_AF2122/97 NSVFGGTAPLIYQALKERDQ---VPMFIAYVTACIAVSLIVYVFFIKNKA
Rv3476c|M.tuberculosis_H37Rv NSVFGGTAPLIYQALKERDQ---VPMFIAYVTACIAVSLIVYVFFIKNKA
Mflv_3055|M.gilvum_PYR-GCK WTAN-WVITVSFPELRNFLGA-AYGFYALCAVLSGLFVWKWVRETKGVSL
Mvan_3472|M.vanbaalenii_PYR-1 WAAN-WLITVTFPGLREHLGL-AYGFYGLCAVLSGLFVWRWVMETKGVSL
MAB_0365c|M.abscessus_ATCC_199 LVANAVVSMYFLTLLTALGGVVAFGVFGVLATAALVFVHVLAPETKGRNL
TH_3400|M.thermoresistible__bu TSLFGGTAPAANDWLIQQTGN-NLMPAFYMMAACLVGMLALVKLPETARC
*
MUL_3187|M.ulcerans_Agy99 EQRPLEELALPLTVV--------DAEDDDEPADG-AGARGAQHPGS----
MMAR_2574|M.marinum_M EQRPLEELALPLTVA--------DAEDDDEPADG-AGARGAQHPGS----
MAV_3053|M.avium_104 EQRPLEDLALPLTVD--------DAEDTEPQGDS-APVD-----------
MSMEG_3536|M.smegmatis_MC2_155 EGQKLEDLAMPLTADEGDGESGGDAENADDQADERARARKEREARIAARV
Mb3503c|M.bovis_AF2122/97 DTYLDREQGFAFYGHA----------------------------------
Rv3476c|M.tuberculosis_H37Rv DTYLDREQGFAFYGHA----------------------------------
Mflv_3055|M.gilvum_PYR-GCK EDMHGEILHGDKSATG----------------------------------
Mvan_3472|M.vanbaalenii_PYR-1 EDMHGEILHADKTATG----------------------------------
MAB_0365c|M.abscessus_ATCC_199 EEIRHFWESGGTWPET----------------------------------
TH_3400|M.thermoresistible__bu PIGGTEIPGTEDAPPPVEYDLVGNR-------------------------
MUL_3187|M.ulcerans_Agy99 --------------------------------------------------
MMAR_2574|M.marinum_M --------------------------------------------------
MAV_3053|M.avium_104 --------------------------------------------------
MSMEG_3536|M.smegmatis_MC2_155 ERQRAAQSRARRYRPGPPPGWEGAWREPPTPGVPESALDHEIDAIARAVA
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3055|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3472|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0365c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------------------
MUL_3187|M.ulcerans_Agy99 --------------------------------------------------
MMAR_2574|M.marinum_M --------------------------------------------------
MAV_3053|M.avium_104 --------------------------------------------------
MSMEG_3536|M.smegmatis_MC2_155 DNESMTVRELHRAVGAQYWGPGEFRKALREALAENRIARKARGRVGPPSG
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3055|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3472|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0365c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------------------
MUL_3187|M.ulcerans_Agy99 ---------
MMAR_2574|M.marinum_M ---------
MAV_3053|M.avium_104 ---------
MSMEG_3536|M.smegmatis_MC2_155 MAEPGTPNR
Mb3503c|M.bovis_AF2122/97 ---------
Rv3476c|M.tuberculosis_H37Rv ---------
Mflv_3055|M.gilvum_PYR-GCK ---------
Mvan_3472|M.vanbaalenii_PYR-1 ---------
MAB_0365c|M.abscessus_ATCC_199 ---------
TH_3400|M.thermoresistible__bu ---------