For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QRGILVALTAASVGLIYGYDLSIIAGAQLFITEEFGLSTHQQELLTTMVVIGQIVGALGAGVLANAIGRK KSVVMLLVAYTMFAVLGALSVSLPMLLAARFLLGLAVGVSIVVVPVYVAESAPAAVRGSLLTVYQLTTVS GLIVGYLTGYLLAGTHSWRWMLGLATVPAMLLLPLLIRMPDTPRWYVMKGRIQEARAALLRVDPAADVEE ELAEIGTALSEGSGGVSEMLRPPFLRATIFVITLGFLIQITGINAIIYYSPRIFEAMGFTGDFALLGLPA LVQIAGVAAVITSLLLVDRVGRRPILLSGIAIMIVADVALMAVFARGQGAAILGFAGILLFIIGYTMGFG SLGWVYASESFPTRLRSIGSSTMLTANLIANAIVAAVFLTMLHSLGGSGAFAVFGVLALVAFGVVYRYAP ETKGRQLEEIRHFWENGGRWPEKLTEAP*
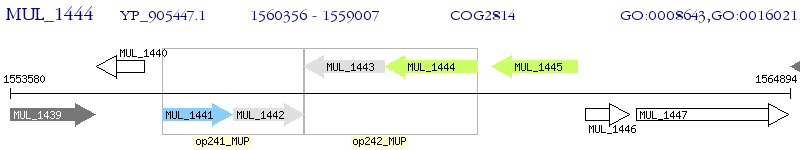
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1444 | sugI | - | 100% (449) | sugar-transport integral membrane protein SugI |
| M. ulcerans Agy99 | MUL_3187 | - | 3e-14 | 22.54% (426) | sugar transporter |
| M. ulcerans Agy99 | MUL_0949 | - | 8e-13 | 25.81% (434) | integral membrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3364 | sugI | 1e-178 | 68.89% (450) | sugar-transport integral membrane protein SugI |
| M. gilvum PYR-GCK | Mflv_3055 | - | 9e-52 | 28.67% (457) | sugar transporter |
| M. tuberculosis H37Rv | Rv3331 | sugI | 1e-178 | 68.89% (450) | sugar-transport integral membrane protein SugI |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0365c | - | 1e-144 | 58.89% (450) | sugar-transport integral membrane protein |
| M. marinum M | MMAR_1191 | sugI | 0.0 | 99.55% (449) | sugar-transport integral membrane protein SugI |
| M. avium 104 | MAV_4306 | - | 0.0 | 77.48% (444) | transporter, major facilitator superfamily protein |
| M. smegmatis MC2 155 | MSMEG_5559 | - | 2e-59 | 32.11% (436) | metabolite/sugar transport protein |
| M. thermoresistible (build 8) | TH_2640 | - | 3e-30 | 28.26% (322) | PUTATIVE sugar transporter |
| M. vanbaalenii PYR-1 | Mvan_3472 | - | 8e-51 | 28.70% (453) | sugar transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3364|M.bovis_AF2122/97 MTTLWQPHRNDYSPIPGRGVHARRGARRPRPRGGRAERPGTGQLTRSGRR
Rv3331|M.tuberculosis_H37Rv MTTLWQPHRNDYSPIPGRGVHARRGARRPRPRGGRAERPGTGQLTRSGRR
MUL_1444|M.ulcerans_Agy99 -----------------------------------------------MQR
MMAR_1191|M.marinum_M -----------------------------------------------MQR
MAV_4306|M.avium_104 -------------------------------------------MARGSRR
MAB_0365c|M.abscessus_ATCC_199 -------------------------------------MSLITDLRSTSRL
MSMEG_5559|M.smegmatis_MC2_155 ----------------------------MSTDTQDSADARAKTPGQLTGA
Mflv_3055|M.gilvum_PYR-GCK ------------------------MAGQGGPAGESPELYEKGQDFSSGKT
Mvan_3472|M.vanbaalenii_PYR-1 ------------------------MAGQGGPVGDGPEIFEQGQEFSSGKT
TH_2640|M.thermoresistible__bu --------------------------------------------------
Mb3364|M.bovis_AF2122/97 ALLVGLTAASVGVLYGYDLSAIAGALLSLSEEFELTTREQELLTTTAVLG
Rv3331|M.tuberculosis_H37Rv ALLVGLTAASVGVLYGYDLSAIAGALLSLSEEFELTTREQELLTTTAVLG
MUL_1444|M.ulcerans_Agy99 GILVALTAASVGLIYGYDLSIIAGAQLFITEEFGLSTHQQELLTTMVVIG
MMAR_1191|M.marinum_M GILVALTAASVGLIYGYDLSIIAGAQLFITEEFGLSTHQQELLTTMVVIG
MAV_4306|M.avium_104 GLLVGLTAASVGVIYGYDLSIIAGAQLFVTEDFGLSTRQQELLTTMAVIG
MAB_0365c|M.abscessus_ATCC_199 GVMVAVTAAAVGVIYGYDSSNIAGALLFLTEDLHLSTHDQQVAATAVVIG
MSMEG_5559|M.smegmatis_MC2_155 VVIVALVSAVSGMLYGYDTGIISGALLQITKDFSIAEAWKQVIAASILLG
Mflv_3055|M.gilvum_PYR-GCK AIRIASVAALGGLLFGYDSAVINGAVASIQEDFGIGNYALGLAVASALLG
Mvan_3472|M.vanbaalenii_PYR-1 AVRIASVAALGGLLFGYDSAVINGAVDSIQEDFGIGNAELGFAVASALLG
TH_2640|M.thermoresistible__bu --------------------------------------------------
Mb3364|M.bovis_AF2122/97 QIAGALGGGILANAIGRKKSVVLIVAGYAVFALLGATSVSVPMLVVARLL
Rv3331|M.tuberculosis_H37Rv QIAGALGGGILANAIGRKKSVVLIVAGYAVFALLGATSVSVPMLVVARLL
MUL_1444|M.ulcerans_Agy99 QIVGALGAGVLANAIGRKKSVVMLLVAYTMFAVLGALSVSLPMLLAARFL
MMAR_1191|M.marinum_M QIVGALGAGVLANAIGRKKSVVMLLVAYTMFAVLGSLSVSLPMLLAARFL
MAV_4306|M.avium_104 QIGGALFAGVLANAIGRRRSVLLILSGYAVFALLAAFSVGLPMLLTARLL
MAB_0365c|M.abscessus_ATCC_199 EIFGALIGGPLSNKIGRKRSMVLVAATFGIFSLLSALAVDLNTLAAARFL
MSMEG_5559|M.smegmatis_MC2_155 AVIGALVCSHLSQKRGRKGTLLMLAVVFVIGSLWCAISPNPVLLSVGRLV
Mflv_3055|M.gilvum_PYR-GCK AAVGALSAGRIADRIGRIAVMKIAAVLFLLSAFGTAFAPETITLVIGRIV
Mvan_3472|M.vanbaalenii_PYR-1 AAAGAMTAGRIADRIGRIAVMKIAAVLFLVSAFGTGFAHEVWAVVLFRIV
TH_2640|M.thermoresistible__bu --------------------------------------------------
Mb3364|M.bovis_AF2122/97 LGVTIGLSVVVVPVYVAESAPAAVRGSLVTAYQLATLSGIVVGYLVGYLL
Rv3331|M.tuberculosis_H37Rv LGVTIGLSVVVVPVYVAESAPAAVRGSLVTAYQLATLSGIVVGYLVGYLL
MUL_1444|M.ulcerans_Agy99 LGLAVGVSIVVVPVYVAESAPAAVRGSLLTVYQLTTVSGLIVGYLTGYLL
MMAR_1191|M.marinum_M LGLAVGVSIVVVPVYVAESAPAAVRGSLLTVYQLTTVSGLIVGYLTGYLL
MAV_4306|M.avium_104 LGLTIGVTVVVVPVYVAESAPTAVRGALLTAYQLAIVSGLIVGYLSGYLL
MAB_0365c|M.abscessus_ATCC_199 LGLTVGVSVVVVPVFVAESSPTKIRGSMLVLYQLACVTGIIAGYLIAWAL
MSMEG_5559|M.smegmatis_MC2_155 LGFAVGGATQTAPMYVAELSPPKYRGRLVLCFQIAIGVGIVIATIVG--A
Mflv_3055|M.gilvum_PYR-GCK GGVGVGVASVIAPAYIAETSPPGIRGRLGSLQQLAIVSGIFASFAVNYLL
Mvan_3472|M.vanbaalenii_PYR-1 GGIGVGVASVIAPAYIAETSPPGIRGRLGSLQQLAIVSGIFASFAVNWLL
TH_2640|M.thermoresistible__bu ---------------------------------LAIVSGIFLSLLIDGIL
:: *:. .
Mb3364|M.bovis_AF2122/97 AGSHG------------WRAMFGLAAAPATLLLPLLWRMPDTARWYLLKG
Rv3331|M.tuberculosis_H37Rv AGSHG------------WRAMFGLAAAPATLLLPLLWRMPDTARWYLLKG
MUL_1444|M.ulcerans_Agy99 AGTHS------------WRWMLGLATVPAMLLLPLLIRMPDTPRWYVMKG
MMAR_1191|M.marinum_M AGTHS------------WRWMLGLATVPAMLLLPLLIRMPDTPRWYVMKG
MAV_4306|M.avium_104 AGTHS------------WRWMLGLACVPAVLLLPLVFRMPDTARWYLLKG
MAB_0365c|M.abscessus_ATCC_199 SSTGS------------WRLMLGLAAVPAALVLIALLKLPDTARWYMMRG
MSMEG_5559|M.smegmatis_MC2_155 SEHIP------------WRWSIGAAAVPAAIMLVLLLRLPESPRWLIKDG
Mflv_3055|M.gilvum_PYR-GCK QWLAGGPNEPLWLGMDAWRWMFLAMAVPAVVYGGLAFTIPESPRYLVATH
Mvan_3472|M.vanbaalenii_PYR-1 QWAAGGPNEPLWFGLDAWRWMFLAMALPAVLYGVLAFTIPESPRYLVASH
TH_2640|M.thermoresistible__bu AALAGGSREELWLNMEAWRWMFLMMAVPAVLYGALTFTIPESPRYLVATH
** : ** : :*::.*: :
Mb3364|M.bovis_AF2122/97 RIADARSALRRIQP-EADIDAELADMAAAV-DERGG----GIGEMVRRP-
Rv3331|M.tuberculosis_H37Rv RIADARSALRRIQP-EADIDAELADMAAAV-DERGG----GIGEMVRRP-
MUL_1444|M.ulcerans_Agy99 RIQEARAALLRVDP-AADVEEELAEIGTAL-SEGSG----GVSEMLRPP-
MMAR_1191|M.marinum_M RIQEARAALLRVDP-AADVEEELAEIGTAL-SEGSG----GVSEMLRPP-
MAV_4306|M.avium_104 RVDDARRALLRVEP-AARVDDELAEIGRAV-SEEAASLPAMLAEMVRSP-
MAB_0365c|M.abscessus_ATCC_199 DRARAREVLSMVDR-DVDIDHELDEIAEALRAEGGGSVRARLREMVTAP-
MSMEG_5559|M.smegmatis_MC2_155 DPDKAREVLERVRPDGYDIDGELDEMTTLVRKEQTAKTR-GWPGLRAAW-
Mflv_3055|M.gilvum_PYR-GCK KIPEARRVLSMLLG-QKNLEITITRIRETLEREDRP----SWRDLKKPTG
Mvan_3472|M.vanbaalenii_PYR-1 KIPEARRVLSMLLG-KKNLEITITRIRETLEREDKP----SWRDMRKPTG
TH_2640|M.thermoresistible__bu RVPEARRVLSRLLG-AKNLEITINRIERSLRAEKPP----SWSDLRKPTG
** .* : :: : : : * :
Mb3364|M.bovis_AF2122/97 -YLRATLFVIALGFLVQITGINAIIYYSPRLFAAMGFAGYFAMLALPAMV
Rv3331|M.tuberculosis_H37Rv -YLRATLFVIALGFLVQITGINAIIYYSPRLFAAMGFAGYFAMLALPAMV
MUL_1444|M.ulcerans_Agy99 -FLRATIFVITLGFLIQITGINAIIYYSPRIFEAMGFTGDFALLGLPALV
MMAR_1191|M.marinum_M -FLRATIFVITLGFLIQITGINAIIYYSPRIFEAMGFTGDFALLGLPALV
MAV_4306|M.avium_104 -YRRATVFVVVLGFLIQITGINAIIYYSPRIFEAMGFTGNFALLALPALV
MAB_0365c|M.abscessus_ATCC_199 -YRRATFFVVGLGFFIQITGINAVVYYGPRIFEAMGMSGYFAKLGLPALV
MSMEG_5559|M.smegmatis_MC2_155 -VRPALILGCGIAIFTQLSGIEMIIYYSPTILTDNGFS-ESVALQVSVAL
Mflv_3055|M.gilvum_PYR-GCK GIYGIVWVGLGLSIFQQFVGINVIFYYSNVLWQAVGFSADESAVYT-VIT
Mvan_3472|M.vanbaalenii_PYR-1 GLYGIVWVGLGLSIFQQFVGINVIFYYSNVLWQAVGFSADESAVYT-VIT
TH_2640|M.thermoresistible__bu GMYGIVWVGLGLSIFQQFVGINVIFYYSNVLWEAVGFDESQSFLIT-VIT
. :.:: *: **: :.**. : *: : .
Mb3364|M.bovis_AF2122/97 QVAGLAAVCASLFLVDRLGRRPILLSGIATMITADAVLITVFANDSD---
Rv3331|M.tuberculosis_H37Rv QVAGLAAVCASLFLVDRLGRRPILLSGIATMITADAVLITVFANDSD---
MUL_1444|M.ulcerans_Agy99 QIAGVAAVITSLLLVDRVGRRPILLSGIAIMIVADVALMAVFAR------
MMAR_1191|M.marinum_M QIAGVAAVITSLLLVDRVGRRPILLSGIAIMIVADVALMAVFAR------
MAV_4306|M.avium_104 QVAGLVAVGTALLLVDRVGRRPILLCGTAMMIVADVVLVAVFGR------
MAB_0365c|M.abscessus_ATCC_199 QVAALLAVFISMSSIDRMGRRPILMIGIGIMIVADALLVGVFAVGGSS--
MSMEG_5559|M.smegmatis_MC2_155 GVSYLVAQLVGLSIIDKVGRRRLTLIMIPGAAVSLFVLGTLFATGHS---
Mflv_3055|M.gilvum_PYR-GCK SVVNVLTTLIAIALIDKIGRKPLLLIGSVGMAVTLSTMAFIFANAELIEN
Mvan_3472|M.vanbaalenii_PYR-1 SVINVLTTLIAIALIDKIGRKPLLLIGSSGMAVTLITMAVIFANATVGAD
TH_2640|M.thermoresistible__bu SVTNIVTTLIAIALIDKIGRKPLLLIGSTGMAGTLGVMAVIFGTAPVIDG
: : : .: :*::**: : : : : :*.
Mb3364|M.bovis_AF2122/97 ---------GGTGLVLGFAGVLLFIIGFNFGFGSLVWVYAAESFPSRLRS
Rv3331|M.tuberculosis_H37Rv ---------GGTGLVLGFAGVLLFIIGFNFGFGSLVWVYAAESFPSRLRS
MUL_1444|M.ulcerans_Agy99 ---------GQGAAILGFAGILLFIIGYTMGFGSLGWVYASESFPTRLRS
MMAR_1191|M.marinum_M ---------GQGAAILGFAGILLFIIGYTMGFGSLGWVYASESFPTRLRS
MAV_4306|M.avium_104 ---------GPGGVIAGFAGVLLFIFGYTMGFGSLGWVYASESFPSRLRS
MAB_0365c|M.abscessus_ATCC_199 --------FGGLLTFLGFLGIVLIAVGFTFGFGSLVWVYAGESFPARLRS
MSMEG_5559|M.smegmatis_MC2_155 ---------GRDSVPFIVACLVVFMLFNAGGLQLMGWLTGSETYPLAVRP
Mflv_3055|M.gilvum_PYR-GCK EAGEMVPSLPGASGVIALIAANLFVVAFGMSWGPVVWVLLGEMFPNRIRA
Mvan_3472|M.vanbaalenii_PYR-1 GN----PSLPGASGVIALIAANLFVVAFGMSWGPVVWVLLGEMFPNRIRA
TH_2640|M.thermoresistible__bu QP-----QLGDVAGPVALVAANLFVVSFGMSWGPVVWVLLGEMFPNRIRA
. :: . . : *: .* :* :*.
Mb3364|M.bovis_AF2122/97 MGSSLMLTSTLTANAIVAAFSLTMLRVLGGAGVFAVFGTFAVVAFVVVYR
Rv3331|M.tuberculosis_H37Rv MGSSPMLTSTLTANAIVAAFSLTMLRVLGGAGVFAVFGTFAVVAFVVVYR
MUL_1444|M.ulcerans_Agy99 IGSSTMLTANLIANAIVAAVFLTMLHSLGGSGAFAVFGVLALVAFGVVYR
MMAR_1191|M.marinum_M IGSSTMLTANLIANAIVAGVFLTMLHSLGGSGAFAVFGVLALVAFGVVYR
MAV_4306|M.avium_104 IGSSTMLTSNLVANAIVAAVFLTLLHSLGGAGTFAVFAVLAVVAFAFVHR
MAB_0365c|M.abscessus_ATCC_199 YGASAMLTSDLVANAVVSMYFLTLLTALGGVVAFGVFGVLATAALVFVHV
MSMEG_5559|M.smegmatis_MC2_155 AGTAAQSAALWGTNLLITLTLLTLISGIGVGPAMWLYGLFNVAAWLFVYF
Mflv_3055|M.gilvum_PYR-GCK AALGLAAAGQWTANWVITVSFPELRNFLG--AAYGFYALCAVLSGLFVWK
Mvan_3472|M.vanbaalenii_PYR-1 AALGLAAAGQWAANWLITVTFPGLREHLG--LAYGFYGLCAVLSGLFVWR
TH_2640|M.thermoresistible__bu AALGLAAAGQWTANWVITVTFPGLREHLG--PAYGFYALCAVLSLLFVWR
. . :. :* ::: : :* . .:. : .*
Mb3364|M.bovis_AF2122/97 FAPETKGRKLEEIRHFWENGGRWPAERSPAADEP
Rv3331|M.tuberculosis_H37Rv FAPETKGRKLEEIRHFWENGGRWPAERSPAADEP
MUL_1444|M.ulcerans_Agy99 YAPETKGRQLEEIRHFWENGGRWPEKLTEAP---
MMAR_1191|M.marinum_M YAPETKGRQLEEIRHFWENGGRWPEKLTEAP---
MAV_4306|M.avium_104 YAPETKGRQLEDIRHFWENGGRWD----------
MAB_0365c|M.abscessus_ATCC_199 LAPETKGRNLEEIRHFWESGGTWPET--------
MSMEG_5559|M.smegmatis_MC2_155 RMPDLTGRSLEEIEGKLRDGKFRPADFAR-----
Mflv_3055|M.gilvum_PYR-GCK WVRETKGVSLEDMHGEILHGDKS-ATG-------
Mvan_3472|M.vanbaalenii_PYR-1 WVMETKGVSLEDMHGEILHADKT-ATG-------
TH_2640|M.thermoresistible__bu WVAETKGRTLEDMQPDVPGHDRASAPG-------
: .* **::.