For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSARAQTGTISTHVPARLDRLPWGRFHWRVVIGLGGVWVLDGLEVTMVGNVSARLTEHGSGINLTPAQIG VAASFYIAGACLGALFFGHLTDRFGRRNLFLLTLAVYLTATVATAFAFAPWYFFIARFFTGSGIGGEYAA INSAIDELIPARVRGRVDLIINGTYWLGSAAGAAGALVLLDTSNFAPNMGWRLAFGLGAIFGIFVLLVRR NVPESPRWLFIHGREQEAEQIVREIEEEVERDTGQRLPDPPGRPLKIRQRQRISLWEIARVAFTRYPRRA FLGLALFIGQAFLYNGVTFNLGTLLSGFYGVESGHVPLFFIVWALSNFVGPLVLGRLFDTVGRKPMITMS YIGSAVIAVVLAVIFVNQTGGVWTFIIVLAAAFFLASAGASAAYLTVSEIFPMETRALAIAFFYAVGTAI GGISGPLLFGQLIESGQRSQVFWSFIIGAAVMAVAGLVELWLGIAAEQRPLEELALPLTVADAEDDDEPA DGAGARGAQHPGS
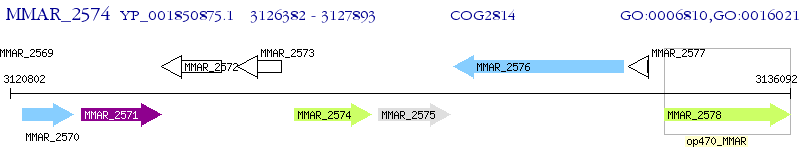
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2574 | - | - | 100% (503) | sugar transporter |
| M. marinum M | MMAR_4238 | - | 2e-16 | 27.02% (396) | integral membrane transport protein |
| M. marinum M | MMAR_1191 | sugI | 6e-14 | 22.05% (449) | sugar-transport integral membrane protein SugI |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3503c | kgtP | 5e-17 | 23.64% (368) | dicarboxylic acid transport integral membrane protein KgtP |
| M. gilvum PYR-GCK | Mflv_3055 | - | 8e-20 | 24.76% (416) | sugar transporter |
| M. tuberculosis H37Rv | Rv3476c | kgtP | 2e-16 | 23.37% (368) | dicarboxylic acid transport integral membrane protein KgtP |
| M. leprae Br4923 | MLBr_01562 | - | 2e-05 | 24.52% (208) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_0365c | - | 7e-20 | 24.94% (437) | sugar-transport integral membrane protein |
| M. avium 104 | MAV_3053 | - | 0.0 | 81.74% (493) | sugar transport protein |
| M. smegmatis MC2 155 | MSMEG_3536 | - | 0.0 | 72.95% (499) | sugar transport protein |
| M. thermoresistible (build 8) | TH_3400 | - | 1e-16 | 24.16% (447) | PUTATIVE General substrate transporter |
| M. ulcerans Agy99 | MUL_3187 | - | 0.0 | 99.79% (473) | sugar transporter |
| M. vanbaalenii PYR-1 | Mvan_3472 | - | 1e-22 | 25.74% (408) | sugar transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3055|M.gilvum_PYR-GCK ---------------MAGQGGPAGESPELYEKGQDFSSGKTAIRIASVAA
Mvan_3472|M.vanbaalenii_PYR-1 ---------------MAGQGGPVGDGPEIFEQGQEFSSGKTAVRIASVAA
MMAR_2574|M.marinum_M -------------MSARAQTGTISTHVPARLDRLPWGRFHWRVVIGLGG-
MUL_3187|M.ulcerans_Agy99 -------------------------------------------MIGLGG-
MAV_3053|M.avium_104 -------------MSAGAQTNTITTHVPARLDRLPWSRFHWRVVIGLGG-
MSMEG_3536|M.smegmatis_MC2_155 -------------MSSGVQTGTITTQVPSRLDRLPWSRFHWRVVLGLGG-
Mb3503c|M.bovis_AF2122/97 --------------------MTVSIAPPSRPSQAETRRAIWNTIRGSSGN
Rv3476c|M.tuberculosis_H37Rv --------------------MTVSIAPPSRPSQAETRRAIWNTIRGSSGN
MAB_0365c|M.abscessus_ATCC_199 ----------------------------MSLITDLRSTSRLGVMVAVTAA
TH_3400|M.thermoresistible__bu ----------------LTTERTDPPQDPSVEQEQPPGVLRKAIAASAVGN
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Mflv_3055|M.gilvum_PYR-GCK LGGLLFGYDSAV----------INGAVASIQEDFG---IGNYALGLAVAS
Mvan_3472|M.vanbaalenii_PYR-1 LGGLLFGYDSAV----------INGAVDSIQEDFG---IGNAELGFAVAS
MMAR_2574|M.marinum_M -VWVLDGLEVTM----------VGNVSARLTEHGSGINLTPAQIGVAASF
MUL_3187|M.ulcerans_Agy99 -VWVLDGLEVTM----------VGNVSARLTEHGSGINLTPAQIGVAASF
MAV_3053|M.avium_104 -VWVLDGLEVTM----------VGNVSARLMEPGSGIALNPAQIGMAAAI
MSMEG_3536|M.smegmatis_MC2_155 -VWILDGLEVTM----------VGNVASRLTESGSGIELSAGQIGIAAAV
Mb3503c|M.bovis_AF2122/97 LVEWYDVYVYTV----------FATYFEDQFFDRADRNSTVYVYAIFAVT
Rv3476c|M.tuberculosis_H37Rv LVEWYDVYVYTV----------FATYFEDQFFDRADRNSTVYVYAIFAVT
MAB_0365c|M.abscessus_ATCC_199 AVGVIYGYDSSN----------IAGALLFLTEDLH---LSTHDQQVAATA
TH_3400|M.thermoresistible__bu ATEWFDYGLYAY----------GVTYISVAIFPGDGATATLLALMTFAVS
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
Mflv_3055|M.gilvum_PYR-GCK ALLGAAVGALSAGRIADRIGRIAVMKIAAVLFLLSAFGTAFAP-------
Mvan_3472|M.vanbaalenii_PYR-1 ALLGAAAGAMTAGRIADRIGRIAVMKIAAVLFLVSAFGTGFAH-------
MMAR_2574|M.marinum_M YIAGACLGALFFGHLTDRFGRRNLFLLTLAVYLTATVATAFA--------
MUL_3187|M.ulcerans_Agy99 YIAGACLGALFFGHLTDRFGRRNLFLLTLAVYLTATVATAFA--------
MAV_3053|M.avium_104 YIAGACSGALFFGHLTDRFGRRNLFILTLALYLIATVATAFA--------
MSMEG_3536|M.smegmatis_MC2_155 YITGACLGALFFGQLTDRFGRKKLFLLTLAVYLVATVATAFA--------
Mb3503c|M.bovis_AF2122/97 FVT-RPVGSWFFGRFADRRGRRAALTFSVSLMAACSLIVALVPSRSSIGV
Rv3476c|M.tuberculosis_H37Rv FVT-RPVGSWFLGRFADRRGRRAALTFSVSLMAACSLIVALVPSRSSIGV
MAB_0365c|M.abscessus_ATCC_199 VVIGEIFGALIGGPLSNKIGRKRSMVLVAATFGIFSLLSALAV-------
TH_3400|M.thermoresistible__bu FLV-RPLGGFVWGPLGDRLGRRQVLAITILLMAGATFCVGLVPSYDSIGL
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVA--------
: * : : ** : . :. ...
Mflv_3055|M.gilvum_PYR-GCK -ETITLVIGRIVGGVGVGVASVIAPAYIAETSPPG-IRGRLGSLQQLAIV
Mvan_3472|M.vanbaalenii_PYR-1 -EVWAVVLFRIVGGIGVGVASVIAPAYIAETSPPG-IRGRLGSLQQLAIV
MMAR_2574|M.marinum_M FAPWYFFIARFFTGSGIGGEYAAINSAIDELIPAR-VRGRVDLIINGTYW
MUL_3187|M.ulcerans_Agy99 FAPWYFFIARFFTGSGIGGEYAAINSAIDELIPAR-VRGRVDLIINGTYW
MAV_3053|M.avium_104 FAPWYFFLTRFFTGAGIGGEYAAINSAIDELIPAR-VRGRVDLVINGTYW
MSMEG_3536|M.smegmatis_MC2_155 FAPWFFFVCRFFTGAGIGGEYAAINSAIDELIPAR-VRGRVDLIINGSYW
Mb3503c|M.bovis_AF2122/97 AAPILLILCRLVQGFATGGEYGTSVTYMSEAATRE-RRGYFSSFQYVTLV
Rv3476c|M.tuberculosis_H37Rv AAPILLILCRLVQGFATGGEYGTSATYMSEAATRE-RRGYFSSFQYVTLV
MAB_0365c|M.abscessus_ATCC_199 -DLNTLAAARFLLGLTVGVSVVVVPVFVAESSPTK-IRGSMLVLYQLACV
TH_3400|M.thermoresistible__bu WAPVLMVILRMIQGFSTGGEYGGAATFMAEYAPSR-RRGLLGSFLEFGTL
MLBr_01562|M.leprae_Br4923 WDEVTLVIARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTA
. *:. * . : . *. .
Mflv_3055|M.gilvum_PYR-GCK SGIFASFAVNYLLQWLAGGPNEPLWLGMDAWRWMFLAMAVPAVVYGGLAF
Mvan_3472|M.vanbaalenii_PYR-1 SGIFASFAVNWLLQWAAGGPNEPLWFGLDAWRWMFLAMALPAVLYGVLAF
MMAR_2574|M.marinum_M LGSAAGAAGALVLLDTSN------FAPNMGWRLAFGLGAIFGIFVLLVRR
MUL_3187|M.ulcerans_Agy99 LGSAAGAAGALVLLDTSN------FAPNMGWRLAFGLGAIFGIFVLLVRR
MAV_3053|M.avium_104 LGSAAGAGGALILLDTSN------FAADLGWRLAFGIGAILGIFVLLVRR
MSMEG_3536|M.smegmatis_MC2_155 LGAAGGALGALLLLNTAI------FPADIGWRIAFGIGAVFGLVVLVVRR
Mb3503c|M.bovis_AF2122/97 GGHVLAQFTLLVILAVFTRE----QVHEFGWRIGFAVGGGAAIVVFWLRR
Rv3476c|M.tuberculosis_H37Rv GGHVLAQFTLLVILAVFTRE----QVHEFGWRIGFAVGGGAAIVVFWLRR
MAB_0365c|M.abscessus_ATCC_199 TGIIAGYLIAWALSSTGS------------WRLMLGLAAVPAALVLIALL
TH_3400|M.thermoresistible__bu AGFSLGALMMLGVSLMLSDD----QMNTWGWRVPFLIAAPLGLIGLYLRS
MLBr_01562|M.leprae_Br4923 IGSVMGLVVGGALTEVSWR---LAFLVNVPIGLVMIYLARTALHETHKER
* . : : . .
Mflv_3055|M.gilvum_PYR-GCK TIPESPRYLVATHKIPEARRVLSMLLGQKNLEITITRIR------ETLER
Mvan_3472|M.vanbaalenii_PYR-1 TIPESPRYLVASHKIPEARRVLSMLLGKKNLEITITRIR------ETLER
MMAR_2574|M.marinum_M NVPESPRWLFIHGREQEAEQIVREIEEEVERDTGQRLPDPPGRPLKIRQR
MUL_3187|M.ulcerans_Agy99 NVPESPRWLFIHGREQEAEQIVREIEEEVERDTGQRLPDPPGRPLKIRQR
MAV_3053|M.avium_104 NVPESPRWLFIHGREEEAEHIVGEIEEAVQQQTGRPLPEPQGKALRIRQR
MSMEG_3536|M.smegmatis_MC2_155 HVPESPRWLFIHGQEDEAERVVGEIEREVERSTGRPLDEPEG-SLTVRQR
Mb3503c|M.bovis_AF2122/97 TMDES-----------LSQERLTAIKAGRDHDSGS---------------
Rv3476c|M.tuberculosis_H37Rv TMDES-----------LSQERLTAIKAGRDHDSGS---------------
MAB_0365c|M.abscessus_ATCC_199 KLPDTARWYMMRGDRARAREVLSMVDRDVDIDHELDEIAEALR----AEG
TH_3400|M.thermoresistible__bu RLDDTPIFRELQESDREEKSIWTEFKD-----------------------
MLBr_01562|M.leprae_Br4923 MKLDAAGAILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFA
::
Mflv_3055|M.gilvum_PYR-GCK EDRPSWRDLKKPTGGIYGIVWVGLGLSIFQQ-FVGINVIFYYSNVLWQAV
Mvan_3472|M.vanbaalenii_PYR-1 EDKPSWRDMRKPTGGLYGIVWVGLGLSIFQQ-FVGINVIFYYSNVLWQAV
MMAR_2574|M.marinum_M QRISLWEIARVAFTRYPRRAFLGLALFIGQA-FLYNGVTFNLGTLLSGFY
MUL_3187|M.ulcerans_Agy99 QRISLWEIARVAFTRYPRRAFLGLALFIGQA-FLYNGVTFNLGTLLSGFY
MAV_3053|M.avium_104 TAISFREIAAVAFKLYPRRAVLGLALFIGQA-FLYNGVTFNLGTLLSQFY
MSMEG_3536|M.smegmatis_MC2_155 KTISFVEIAKVAFTKYPKRAVLGLALFIGQA-FIYNAFTFNLGTLLSGFF
Mb3503c|M.bovis_AF2122/97 --------LRELATHYWKPLLLCFLVTLGGT-VAFYTYSVNAPAIVKSVY
Rv3476c|M.tuberculosis_H37Rv --------LRELATHYWKPLLLCFLVTLGGT-VAFYTYSVNAPAIVKSVY
MAB_0365c|M.abscessus_ATCC_199 GGSVRARLREMVTAPYRRATFFVVGLGFFIQ-ITGINAVVYYGPRIFEAM
TH_3400|M.thermoresistible__bu -----------LLVEYRGQILRLGGLVVALN-VVNYTLLSYMPTYLQTSI
MLBr_01562|M.leprae_Br4923 VVERTAENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDIL
: . :
Mflv_3055|M.gilvum_PYR-GCK GFSADESAV-YTVITSVVNVLTTLIAIALIDKIGRKPLLLIGSVGMAVTL
Mvan_3472|M.vanbaalenii_PYR-1 GFSADESAV-YTVITSVINVLTTLIAIALIDKIGRKPLLLIGSSGMAVTL
MMAR_2574|M.marinum_M GVESGHVPL-FFIVWALSNFVGPLVLGRLFDTVGRKPMITMSYIGSAVIA
MUL_3187|M.ulcerans_Agy99 GVESGHVPL-FFIVWALSNFVGPLVLGRLFDTVGRKPMITMSYIGSAVIA
MAV_3053|M.avium_104 AVPSGMVPV-FFVLWALSNFAGPLLLGHLFDTVGRKQMITLTYIGSAVVV
MSMEG_3536|M.smegmatis_MC2_155 DVASGAVPI-FYAVWALSNFAGPILLGRLFDTVGRKPMIALAYLGSAAVA
Mb3503c|M.bovis_AF2122/97 GSQAMTATW-INLVGLILLMMLQPIGGMISDKIGRKPLLLWFGVG-GLIY
Rv3476c|M.tuberculosis_H37Rv GSQAMTATW-INLVGLILLMMLQPIGGMISDKIGRKPLLLWFGVG-GLIY
MAB_0365c|M.abscessus_ATCC_199 GMSGYFAKLGLPALVQVAALLAVFISMSSIDRMGRRPILMIGIGIMIVAD
TH_3400|M.thermoresistible__bu GLSTDTALV-VPIIGMLSMMVFLPFAGLLSDRFGRKPMWWISLGGLLALV
MLBr_01562|M.leprae_Br4923 GYSALRAGI-GFIPFVIAMGIGLGVSSQLVSRFSPR-VLTIGGGWLLMVA
: . . .. : :
Mflv_3055|M.gilvum_PYR-GCK STMAFIFANAELIENEAGEMVPSLPGASGVIALIAANLFVVA----FGMS
Mvan_3472|M.vanbaalenii_PYR-1 ITMAVIFANATVGADGN----PSLPGASGVIALIAANLFVVA----FGMS
MMAR_2574|M.marinum_M VVLAVIFVNQTG-------------GVWTFIIVLAAAFFLAS----AGAS
MUL_3187|M.ulcerans_Agy99 VVLAVIFVNQTG-------------GVWTFIIVLAAAFFLAS----AGAS
MAV_3053|M.avium_104 VALALVFLTQAG-------------GVWAFIGVLIVAFFLAS----AGAS
MSMEG_3536|M.smegmatis_MC2_155 VILTVLFVNEIG-------------GVWLFMLVLGVCFFLAS----SGAS
Mb3503c|M.bovis_AF2122/97 TYVLVTYLPETR-------------SPTMSFLLVAVGYVILT----GYCS
Rv3476c|M.tuberculosis_H37Rv TYVLVTYLPETR-------------SPTMSFLLVAVGYVILT----GYCS
MAB_0365c|M.abscessus_ATCC_199 ALLVGVFAVGGSSFG----------GLLTFLGFLGIVLIAVG----FTFG
TH_3400|M.thermoresistible__bu VPMFMLMSTSVP-------------GAIIGFAVLGLLYVPQL----ATIS
MLBr_01562|M.leprae_Br4923 MLYGWAFMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIG
. : .: .
Mflv_3055|M.gilvum_PYR-GCK WGPVVWVLLGEMFPNRIRAAALGLAAAGQWTAN-WVITVSFPELRNFLGA
Mvan_3472|M.vanbaalenii_PYR-1 WGPVVWVLLGEMFPNRIRAAALGLAAAGQWAAN-WLITVTFPGLREHLGL
MMAR_2574|M.marinum_M ---AAYLTVSEIFPMETRALAIAFFYAVGTAIGGISGPLLFGQLIESGQR
MUL_3187|M.ulcerans_Agy99 ---AAYLTVSEIFPMETRALAIAFFYAVGTAIGGISGPLLFGQLIESGQR
MAV_3053|M.avium_104 ---AAYLTVSEIFPMETRALAIAFFYAMGTAIGGITGPLLFGQLIDSGQR
MSMEG_3536|M.smegmatis_MC2_155 ---AAYLTVSEIFPMETRALAIAFFYAIGTAVGGITGPLLFGQLIESGER
Mb3503c|M.bovis_AF2122/97 ---INALVKSELFPAHVRALGVGVGYALANSVFGGTAPLIYQALKERDQ-
Rv3476c|M.tuberculosis_H37Rv ---INALVKSELFPAHVRALGVGVGYALANSVFGGTAPLIYQALKERDQ-
MAB_0365c|M.abscessus_ATCC_199 FGSLVWVYAGESFPARLRSYGASAMLTSDLVANAVVSMYFLTLLTALGGV
TH_3400|M.thermoresistible__bu ------ATFPAMFPTHVRYAGFAIAYNVSTSLFGGTAPAANDWLIQQTGN
MLBr_01562|M.leprae_Br4923 PVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRAL
.
Mflv_3055|M.gilvum_PYR-GCK -AYGFYALCAVLSGLFVWKWVRETKGVSLEDMHGEILHGDKSATG-----
Mvan_3472|M.vanbaalenii_PYR-1 -AYGFYGLCAVLSGLFVWRWVMETKGVSLEDMHGEILHADKTATG-----
MMAR_2574|M.marinum_M -SQVFWSFIIGAAVMAVAGLVELWLGIAAEQRPLEELALPLTVA------
MUL_3187|M.ulcerans_Agy99 -SQVFWSFIIGAAVMAVAGLVELWLGIAAEQRPLEELALPLTVV------
MAV_3053|M.avium_104 -DHVVWSFLIGAVVMAAAGLVELWLGIAAEQRPLEDLALPLTVD------
MSMEG_3536|M.smegmatis_MC2_155 -GLVAVSFLIGAAVMAVGGVVELILGVKAEGQKLEDLAMPLTADEGDGES
Mb3503c|M.bovis_AF2122/97 --VPMFIAYVTACIAVSLIVYVFFIKNKADTYLDREQGFAFYGHA-----
Rv3476c|M.tuberculosis_H37Rv --VPMFIAYVTACIAVSLIVYVFFIKNKADTYLDREQGFAFYGHA-----
MAB_0365c|M.abscessus_ATCC_199 VAFGVFGVLATAALVFVHVLAPETKGRNLEEIRHFWESGGTWPET-----
TH_3400|M.thermoresistible__bu -NLMPAFYMMAACLVGMLALVKLPETARCPIGGTEIPGTEDAPPPVEYDL
MLBr_01562|M.leprae_Br4923 DHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL---
Mflv_3055|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3472|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2574|M.marinum_M --DAEDDDEPADG-AGARGAQHPGS-------------------------
MUL_3187|M.ulcerans_Agy99 --DAEDDDEPADG-AGARGAQHPGS-------------------------
MAV_3053|M.avium_104 --DAEDTEPQGDS-APVD--------------------------------
MSMEG_3536|M.smegmatis_MC2_155 GGDAENADDQADERARARKEREARIAARVERQRAAQSRARRYRPGPPPGW
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0365c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3400|M.thermoresistible__bu VGNR----------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mflv_3055|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3472|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2574|M.marinum_M --------------------------------------------------
MUL_3187|M.ulcerans_Agy99 --------------------------------------------------
MAV_3053|M.avium_104 --------------------------------------------------
MSMEG_3536|M.smegmatis_MC2_155 EGAWREPPTPGVPESALDHEIDAIARAVADNESMTVRELHRAVGAQYWGP
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0365c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mflv_3055|M.gilvum_PYR-GCK --------------------------------------
Mvan_3472|M.vanbaalenii_PYR-1 --------------------------------------
MMAR_2574|M.marinum_M --------------------------------------
MUL_3187|M.ulcerans_Agy99 --------------------------------------
MAV_3053|M.avium_104 --------------------------------------
MSMEG_3536|M.smegmatis_MC2_155 GEFRKALREALAENRIARKARGRVGPPSGMAEPGTPNR
Mb3503c|M.bovis_AF2122/97 --------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------
MAB_0365c|M.abscessus_ATCC_199 --------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------