For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPEFRFSFNVFRIDPVHTFIETCRRAEQWGYDAVFAADHLGIAAPFMTLVAAAAATDRMRVGILLLNAPW WNPALLARDIATTDVLTGGRLEVGLGAGHMKWEYDQADIPFPSFTTRTARLRQTIEELASRFAEDGFAQQ AELREAHNIAVLRPTQRRGFGGYGPPLLVGGTGDQVLRLAAQYADTVSIACLYQVKGRPPGTMRLAIVAE IEQRVAYVRRCAGAARHPELHTLIQEVVLTDDRREVAGMLAAKYGNLLTANEILQSPTLLIGTVEQIARQ LRANRERYGFTHYTIPQPHVAAFAPVITVLGDLN
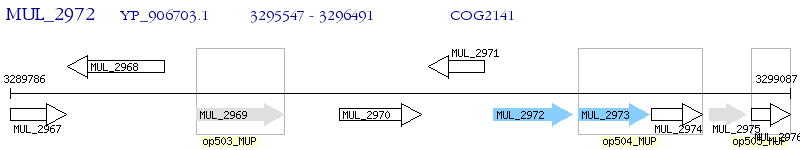
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2972 | - | - | 100% (314) | oxidoreductase |
| M. ulcerans Agy99 | MUL_3024 | - | 6e-20 | 29.29% (280) | oxidoreductase |
| M. ulcerans Agy99 | MUL_3839 | - | 2e-14 | 30.34% (178) | coenzyme F420-dependent oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2917 | - | 3e-35 | 37.59% (274) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1815 | - | 2e-34 | 30.87% (311) | luciferase family protein |
| M. tuberculosis H37Rv | Rv2893 | - | 3e-35 | 37.59% (274) | oxidoreductase |
| M. leprae Br4923 | MLBr_00131 | - | 3e-06 | 24.26% (169) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_2431c | - | 2e-21 | 28.32% (339) | luciferase-like oxidoreductase |
| M. marinum M | MMAR_2775 | - | 1e-179 | 98.41% (314) | oxidoreductase |
| M. avium 104 | MAV_1042 | - | 1e-34 | 31.72% (309) | hypothetical protein MAV_1042 |
| M. smegmatis MC2 155 | MSMEG_5592 | - | 9e-39 | 33.77% (308) | hypothetical protein MSMEG_5592 |
| M. thermoresistible (build 8) | TH_1174 | - | 9e-18 | 29.14% (302) | POSSIBLE OXIDOREDUCTASE |
| M. vanbaalenii PYR-1 | Mvan_0757 | - | 7e-38 | 38.30% (282) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2917|M.bovis_AF2122/97 ------------MTVASTAHHTRRLRFGLAAPLPRAGTQMRAFAQAVEAA
Rv2893|M.tuberculosis_H37Rv ------------MTVASTAHHTRRLRFGLAAPLPRAGTQMRAFAQAVEAA
Mvan_0757|M.vanbaalenii_PYR-1 ------------------------MRFGLTTALPRDGAAAREFAQRVEAA
MUL_2972|M.ulcerans_Agy99 ---------------------MPEFRFSFNVFRIDPVHTFIETCRRAEQW
MMAR_2775|M.marinum_M MTLRLPRTRWLARLPAGSVTPVPEFRFSFNVFRIDPVHTFIETCRRAEQW
Mflv_1815|M.gilvum_PYR-GCK -----MPLPPAVQTWTYDHDVAKEFRFGVGVTRVSTRAKLEDDARRAEAL
MAV_1042|M.avium_104 --------------------MAKDFRFGMSMRFYKSREALLDKAKRAEDA
MSMEG_5592|M.smegmatis_MC2_155 -------------------MMPKEFRFGVSVRNAESQAQVAATARRAEDL
MAB_2431c|M.abscessus_ATCC_199 ---------------MTIRLGYQIPNFSYGTGVAELFPTVVKQVREAEAS
TH_1174|M.thermoresistible__bu ---------------VSIRLGLQIPNFSYGTGVADLFPTVIAQAREADEA
MLBr_00131|M.leprae_Br4923 ---MAGFRFGFVDALVHTLFPPSLPARASIVSGAVLGADSYWVGDHLNAL
. :
Mb2917|M.bovis_AF2122/97 GFDVLAFPDHLVP----------SVSPFAGATAAAMATQR--LHTGTLVL
Rv2893|M.tuberculosis_H37Rv GFDVLAFPDHLVP----------SVSPFAGATAAAMATQR--LHTGTLVL
Mvan_0757|M.vanbaalenii_PYR-1 GIDVLTFADHLAP----------AVAPFSGATAAAAVTSR--LHVGTLVL
MUL_2972|M.ulcerans_Agy99 GYDAVFAADHLG-----------IAAPFMTLVAAAAATDR--MRVGILLL
MMAR_2775|M.marinum_M GYDAVFAADHLG-----------IAAPFMTLVAAAAATDR--MRVGTLVL
Mflv_1815|M.gilvum_PYR-GCK GFDVLHVPDHLG-----------GSAPFPVMTAVAMATTS--LRVGTFVL
MAV_1042|M.avium_104 GFDILCVPDHLG-----------AAAPFPTLTAVAMVTTT--LRLSMYVL
MSMEG_5592|M.smegmatis_MC2_155 GYNVIVVPDHLG-----------APAPFPTLTAIAAATDT--IRVGTCVL
MAB_2431c|M.abscessus_ATCC_199 GFDTVFVMDHFYQLPMIGTPDQPMLEAYTALGALASATST--IQLSTLVT
TH_1174|M.thermoresistible__bu GYDAVFVMDHFYQLPMLGDPDEPMLEAYTALGGLATSTER--VQLGTLVT
MLBr_00131|M.leprae_Br4923 VPRSVATPKYLGVAAKVVPKIDANYEPWTMLGNLAAGNRLNGLRLGVCVT
: .:: .: * . :: . :
Mb2917|M.bovis_AF2122/97 NNDFRHPVDTAREAAGVATLAEGRFELGLGAGHRRSEYDAAGITFDSGAT
Rv2893|M.tuberculosis_H37Rv NNDFRHPVDTAREAAGVATLAEGRFELGLGAGHRRSEYDAAGITFDSGAT
Mvan_0757|M.vanbaalenii_PYR-1 NNDFRHPVETAREAAGVATVSDGRFELGIGAGHMKSEYDSAGIPFDGGGT
MUL_2972|M.ulcerans_Agy99 NAPWWNPALLARDIATTDVLTGGRLEVGLGAGHMKWEYDQADIPFPSFTT
MMAR_2775|M.marinum_M NAPWWNPALLARDIATTDVLTGGRLEVGLGAGHMKWEYDQADIPFPSFTT
Mflv_1815|M.gilvum_PYR-GCK NSAFYRPALLARDVAALNDLSDGRVELGLGTGYVREEFDAAGIPYPTAGE
MAV_1042|M.avium_104 NSAFYKPALLSRDMQALDLLSDGRLEIGLGTGYVREEFEAAEIPYPSARE
MSMEG_5592|M.smegmatis_MC2_155 NACFYKPALLARDVTALRDLSDGRFEVGLGAGYVREEFEAAELPFPSARE
MAB_2431c|M.abscessus_ATCC_199 GNTYRNPALLAKAVTTLDVVSGGRAILGIGAGWFELEHQQLGFEFGTFTD
TH_1174|M.thermoresistible__bu GNTYRNPTLLAKAVTTLDVMSQGRAVLGIGTGWFELEHQQLGYEFGTFTD
MLBr_00131|M.leprae_Br4923 DAGRRNPAVTAQAAATLHLLTRGKAMLGIGVGE-REGNEPYGVEWTKPVA
. .*. :: :: *: :*:*.* . : :
Mb2917|M.bovis_AF2122/97 RVARLIESAHLIRALLDAEPVDFDGQHYRVHAEAG----SLVAPPKVRVP
Rv2893|M.tuberculosis_H37Rv RVARLIESAHLIRALLDAEPVDFDGQHYRVHAEAG----SLVAPPKVRVP
Mvan_0757|M.vanbaalenii_PYR-1 RVSRLVESVTVLRALLHGDAVDFDGDHYRVHAAAG----EIVAAPVHRVP
MUL_2972|M.ulcerans_Agy99 RTARLRQTIEELASRFAEDGFAQQAELREAHNIAVLRPTQRRGFGGYGPP
MMAR_2775|M.marinum_M RTARLRQTIEELASRFAEEGFAQQAELREAHNIAVLRPTQRRGFGGYGPP
Mflv_1815|M.gilvum_PYR-GCK RVDHLKLTAAHIAEHLPDV------------------------------P
MAV_1042|M.avium_104 RVDYLEHMTKYLKEHHPST------------------------------P
MSMEG_5592|M.smegmatis_MC2_155 RIDHLRHTTEYLREHVSDV------------------------------P
MAB_2431c|M.abscessus_ATCC_199 RFERLTEALEIIPPMLRGERPTFEGKYYRTESALN------EPRLRPNIP
TH_1174|M.thermoresistible__bu RFNRLYEALEIIVPMIEGKRPTFEGRYYRVESAAA------EPRFRDRIP
MLBr_00131|M.leprae_Br4923 RFQEALATIRALWDSNGELVSRESQFFPLHNALFD-----LPPYRGKWPE
* :
Mb2917|M.bovis_AF2122/97 LLVGGNGTE-VLRLGGRIAD------IVGLAGISHNRDATQVRFTHFDAD
Rv2893|M.tuberculosis_H37Rv LLVGGNGTE-VLRLGGRIAD------IVGLAGISHNRDATQVRFTHFDAD
Mvan_0757|M.vanbaalenii_PYR-1 ILIGGNGTR-VLQLAGRIAD------IVGFAGISHNRDATRVRLSHFDGA
MUL_2972|M.ulcerans_Agy99 LLVGGTGDQ-VLRLAAQYAD------TVSIACLYQVKGRPPGTMRLAIVA
MMAR_2775|M.marinum_M LLVGGTGDQ-VLRLAAQYAD------TVSIACLYQVKGRPPGTMRLATVA
Mflv_1815|M.gilvum_PYR-GCK IMIAGNGDR-VLRTAARFAD------IVGFTGGDRAATGDE--------D
MAV_1042|M.avium_104 LIIAGNGDR-VLTIAARNAD------IIGLTG--AKVRDVE--------D
MSMEG_5592|M.smegmatis_MC2_155 IMVAGNGDR-LLTVAAQTAD------IVGLTGGDGVPDGDH--------D
MAB_2431c|M.abscessus_ATCC_199 IMIGGSGEKKTFGLAVKYAD------HLNLLSS---------------LD
TH_1174|M.thermoresistible__bu LMIGGSGEKKTIPLAARRFD------HLNLIAG---------------FD
MLBr_00131|M.leprae_Br4923 IWVAAHGPR-MLQATGRYADAWIPIVLVRPTDYSCALEVVRTAASDAGRD
: :.. * . : : * :
Mb2917|M.bovis_AF2122/97 GLADRIAVVR--------HAAGDRFEAIELNALIQAVVCTNDRNA-AAAE
Rv2893|M.tuberculosis_H37Rv GLADRIAVVR--------HAAGDRFEAIELNALIQAVVCTNDRNA-AAAE
Mvan_0757|M.vanbaalenii_PYR-1 GLADRIGVVR--------EAAGERFAQIELNALIQAVVVTTDRQR-AAAE
MUL_2972|M.ulcerans_Agy99 EIEQRVAYVR--------RCAGAARHPELHTLIQEVVLTDDRREV-AGML
MMAR_2775|M.marinum_M EIEQRVAYVR--------RCAGAARHPELHTLIQEVVLTDDRRKV-AGML
Mflv_1815|M.gilvum_PYR-GCK PLGDRIGFVR--------AAAGERFAALELNIAITAIPLDSSGQP-DLTI
MAV_1042|M.avium_104 PFAERVEFVR--------NAAGDRFDSLELNLAITAMPRVGETEP-DLKL
MSMEG_5592|M.smegmatis_MC2_155 PLAERISFLR--------KAADGR--DLELNITFTAIPAAGSEMP-DLRL
MAB_2431c|M.abscessus_ATCC_199 EIPHKLTVLRQRC-----EEAGRDPSTLETSAFFSVFIDDDSATAWKLAA
TH_1174|M.thermoresistible__bu ELPGKVAAVKRSC-----EEIGRDPDTLETSTLVSVMVDDN---------
MLBr_00131|M.leprae_Br4923 PMSITPAAVRGIITGRTRDDVDEALDSVLVRMIALGVPGEAWARHGVEHP
: :: . .
Mb2917|M.bovis_AF2122/97 LAATLGGITPEQ------------VLESPF------LLLGTHEQMAEALA
Rv2893|M.tuberculosis_H37Rv LAATLGGITPEQ------------VLESPF------LLLGTHEQMAEALA
Mvan_0757|M.vanbaalenii_PYR-1 LAAAFG-ADPET------------LLCSPF------VLLGTHEQMAEQLL
MUL_2972|M.ulcerans_Agy99 AAKYGNLLTANE------------ILQSPT------LLIGTVEQIARQLR
MMAR_2775|M.marinum_M AAKYGNLLTANE------------ILQSPT------LLIGTVEQIARQLR
Mflv_1815|M.gilvum_PYR-GCK PRISLPGLSDDE------------LLRHPS------VLSGSTTEIADRIH
MAV_1042|M.avium_104 TRSYSPELTDEQ------------ILSMPS------VLSGSPRDIADTLS
MSMEG_5592|M.smegmatis_MC2_155 TRRFLGHLTDEQ------------LLRHPG------VLAGSASDMADRLR
MAB_2431c|M.abscessus_ATCC_199 SQLSQNGIDLDT------------LPDEARAQVLGRTLVGNPEEIAEQVQ
TH_1174|M.thermoresistible__bu -------ITDDM------------LPEKAR----GRLLVGSAERIAEQLQ
MLBr_00131|M.leprae_Br4923 MGADFAGVQDIIPQTIDEETVVSYAAKVPAALMKEVLFSGTPEEVIDQVA
. : *. : :
Mb2917|M.bovis_AF2122/97 ARQRRFGVSYWTVFDEWAGRASAMRDIAEVIALLRYG------
Rv2893|M.tuberculosis_H37Rv ARQRRFGVSYWTVFDEWAGRASAMRDIAEVIALLRYG------
Mvan_0757|M.vanbaalenii_PYR-1 ARRREFGIGYWTVFDELPGRESALPDISEVIALLR--------
MUL_2972|M.ulcerans_Agy99 ANRERYGFTHYTIPQPH------VAAFAPVITVLGDLN-----
MMAR_2775|M.marinum_M ANRERYGFTHYTIPQPH------VAAFAPVITVLGDLN-----
Mflv_1815|M.gilvum_PYR-GCK GYRDVYGISYVIVQMRH------AEAFGAVIDLLR--------
MAV_1042|M.avium_104 AYRDKYGVSSFTVQDNH------IDNFAKVIAELR--------
MSMEG_5592|M.smegmatis_MC2_155 QLRDTYGVSYFIVQIPH------IETFAKVIAELR--------
MAB_2431c|M.abscessus_ATCC_199 TRVLDQGIDSVTLNAP--ATGHLPGVVELIGNTLRPLFKN---
TH_1174|M.thermoresistible__bu TKVLDAGVDGVIVNMATKVQGYRPGLITALGEALRPRVGA---
MLBr_00131|M.leprae_Br4923 EWR-DHGLKYLVVINGSLVNSSLRKTVSALLPHARVLRGLKKL
*. : . :